- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Saccharification Protocol for Small-scale Lignocellulosic Biomass Samples to Test Processing of Cellulose into Glucose
Published: Vol 6, Iss 1, Jan 5, 2016 DOI: 10.21769/BioProtoc.1701 Views: 12362
Reviewed by: Maria SinetovaHarrie van ErpAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1897 Views

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1248 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 144 Views
Abstract
Second generation biofuels are derived from inedible lignocellulosic biomass of food and non-food crops. Lignocellulosic biomass is mainly composed of cell walls that contain a large proportion of cellulosic and hemicellulosic polysaccharides. An interesting route to generate biofuels and bio-based materials is via enzymatic hydrolysis of cell wall polysaccharides into fermentable sugars, a process called saccharification. The released sugars can then be fermented to fuels, e.g., by use of yeast.
To test the saccharification efficiency of lignocellulosic biomass on a lab-scale, a manual saccharification protocol was established that uses only small amounts of biomass and a low concentration of enzyme. This protocol can be used for different plant species like Arabidopsis thaliana, tobacco, maize and poplar. The low enzyme concentrations make it possible to detect subtle improvements in saccharification yield and to analyze the speed of hydrolysis. Although a specific acid and alkali pretreatment were included, the saccharification step can be preceded by any other pretreatment. Because no advanced equipment is necessary, this protocol can be carried out in many laboratories to analyze saccharification yield. The protocol was initially described in Van Acker et al. (2013).
Materials and Reagents
- pH-indicator paper (pH 1-14) (Merck Millipore Corporation, catalog number: 1109620003 )
- Disposable PD-10 desalting columns (VWR International, catalog number: 95017001 )
- Whatman® qualitative filter paper, Grade 1 (Sigma-Aldrich, catalog number: WHA1001110 )
- Safe-Lock tubes 2 ml (Thermo Fisher Scientific, Eppendorf, catalog number: 3706 )
- Corning® 15 ml centrifuge tubes (Sigma-Aldrich, catalog number: CLS430790 )
- Corning® 50 ml centrifuge tubes (Sigma-Aldrich, catalog number: CLS430290 )
Note: Pricing & availability is not currently available. - Nunc 96-well microplate without lid and flat bottom wells (Thermo Fisher Scientific, catalog number: 269787 )
- Parafilm (Bemis Flexible Packaging)
- Sodium acetate trihydrate (CH3COONa.3H2O) (Sigma-Aldrich, catalog number: S8625-250 G )
- Sodium azide (NaN3) (Sigma-Aldrich, catalog number: 71290-100 g )
- Cellulase from Trichoderma reesei ATCC 26291 (Sigma-Aldrich, catalog number: C2730 )
- Accellerase® BG from Trichoderma reesei (Genencor, DuPont)
- D-glucose (C6H12O6) (Sigma-Aldrich, catalog number: G8270-1 kg )
- Glucose oxidase from Aspergillus niger (Sigma-Aldrich, catalog number: G6125-50KU )
- Peroxidase from horse radish (Roche Diagnostics, catalog number: 10814407001 )
- 2, 2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) (Roche Diagnostics, catalog number: 11112422001 )
- Hydrochloric acid fuming 37% (Merck Millipore Corporation, catalog number: 1003171000 )
- Sodium hydroxide pellets for analysis (Merck Millipore Corporation, catalog number: 1064981000 )
- Ethanol (Merck Millipore Corporation, catalog number: 1009901001 )
- Acetone (Merck Millipore Corporation, catalog number: 1000121000 )
- Glacial acetic acid (Merck Millipore Corporation, catalog number: 1000631000 )
- MilliQ-water
- 0.1 M acetic acid buffer solution (pH 4.5) (see Recipes)
- 0.1 M acetic acid buffer solution (pH 4.8) (see Recipes)
- Glucose oxidase (GOD) – peroxidase (POD) solution (see Recipes)
Equipment
- Balance (Mettler Toledo, model: XP-105 Delta Range)
- Thermoblock (Thermo Fisher Scientific, Eppendorf, model: Thermomixer Compact )
- Temperature controlled benchtop microcentrifuge (Thermo Fisher Scientific, Eppendorf, model: 5417R )
- Temperature controlled microplate spectrophotometer (Molecular Devices, model: Spectra Max 250 )
- Acid-resistant CentriVap Centrifugal Vacuum Concentrator (Labconco, catalog number: 7810016 )
- Oven at 37 °C
- Tripod with clamp and knots (VWR International, catalog numbers: 2410093 and 2410258 )
- Volumetric flask of 100 ml (Duran Group, catalog number: 246712556 )
- Rotilabo® Sealing films for microtest plates (Carl Roth GmbH + Co., catalog number: EN76.1 )
- Microcentrifuge (LabSource, VWR International, catalog number: 37001298 )
Procedure
- Preparing the enzyme mixture
- The enzyme mixture used in this protocol is a mix of cellulase and β-glucosidase (BG). These enzymes first need to be cleared from their stabilizing salts. This is done with a desalting Econo-Pac 10 DG column, fitted with Bio-gel® P-6DG gel, and can be used for both cellulase and BG.
- First remove, by pouring off, the NaN3-solution that prevents growth of fungi on the column.
- Bring 20 ml of acetic acid buffer solution (pH 4.8) on the column and let gravity work.
- Mix 1 ml of enzyme (cellulase or BG) with 2 ml acetic acid buffer solution (pH 4.8) in a Corning® 15 ml centrifuge tube. Bring this mix on the column and let it run through the column. Remove the 3 ml that comes off the column.
- Add 4 ml acetic acid buffer solution (pH 4.8) on the column and collect the 4 ml that comes off the column in a Corning® 15 ml centrifuge tube. This is the desalted enzyme.
- Clean the column by running 100 ml milliQ-water through the column. The washed column can be used again for another desalting.
- Close the column at the bottom and fill the column with 0.02% NaN3. Close the column with a cap on the top and store the column at room temperature.
- When cellulase and BG are both desalted, dilute the desalted BG 350-fold with acetic acid buffer solution (pH 4.8).
- Mix the desalted cellulase and the desalted, diluted BG in a 5:3 ratio.
- Dilute this mix 10-fold with acetic acid buffer solution (pH 4.8). This is the enzyme mixture that will be used in the saccharification assay. It is recommended that the enzyme mixture is used within two months after preparation.
- The enzyme mixture used in this protocol is a mix of cellulase and β-glucosidase (BG). These enzymes first need to be cleared from their stabilizing salts. This is done with a desalting Econo-Pac 10 DG column, fitted with Bio-gel® P-6DG gel, and can be used for both cellulase and BG.
- Preparing GOD-POD solution (Bergmeyer, 1974) and calibration curve
The glucose oxidase (GOD)-peroxidase (POD) method measures indirectly the concentration of glucose (Figure 1). In a first step GOD converts glucose monomers into gluconic acid with the production of hydrogen peroxide. The oxidizing power of hydrogen peroxide is used by POD to oxidize a dye, 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS). Finally, this oxidized form of ABTS will absorb at a wavelength of 405 nm.
Figure 1. Illustration of the reaction mechanism used to measure glucose- Prepare 100 ml of GOD-POD solution as described in recipes. The GOD-POD solution contains 50 mg ABTS, 44.81 mg glucose oxidase and 173 µl of 4% peroxidase. The final volume is adjusted to 100 ml by adding acetic acid buffer solution (pH 4.5).
- Make a dilution series of different glucose concentrations (from 20 µM till 200 µM). Proceed as described in section H and plot as in Figure 2.
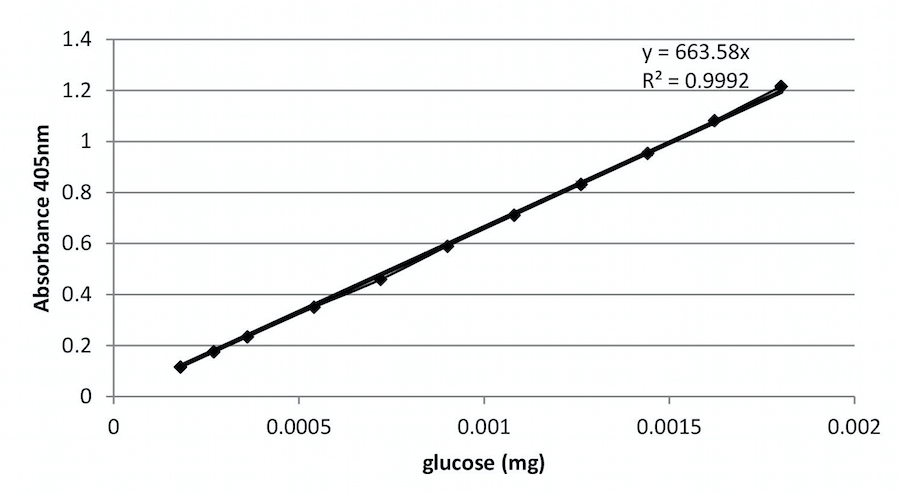
Figure 2. An example of a calibration curve
- Prepare 100 ml of GOD-POD solution as described in recipes. The GOD-POD solution contains 50 mg ABTS, 44.81 mg glucose oxidase and 173 µl of 4% peroxidase. The final volume is adjusted to 100 ml by adding acetic acid buffer solution (pH 4.5).
- Measuring enzyme activity
For publication of saccharification results, it is recommended to report the enzyme activity. As such, the saccharification assay can be repeated with exactly the same conditions. In addition, the number of samples that can be analyzed in one single assay is limited due to practical reasons. To compare saccharification results obtained in different saccharification assays, it is necessary that these assays are performed with the same enzyme activity.- Fill three safe-lock tubes with 4.5 mg of Whatman filter paper. Also include a blank, i.e. a safe-lock tube with no filter paper.
- Add to each safe-lock tube (with or without filter paper) 450 µl acetic acid buffer solution (pH 4.8) and 50 µl of the enzyme mixture.
- Incubate in a thermoblock set at 50 °C for exactly 1 h while shaking at 12.5 Hz.
- Boil the samples and blank for 5 min at 99 °C without shaking.
- Centrifuge at 23,477 x g for 5 min.
- Dilute the supernatants 20 times with acetic acid buffer solution (pH 4.8).
- Measure the glucose release in the diluted supernatant using glucose oxidase-peroxidase (GOD-POD) (see section H) and calculate the activity of the enzyme mixture [expressed in filter paper units (FPU)/ml] according to the formula described in Xiao et al. (2004) and as explained in Figure 3.
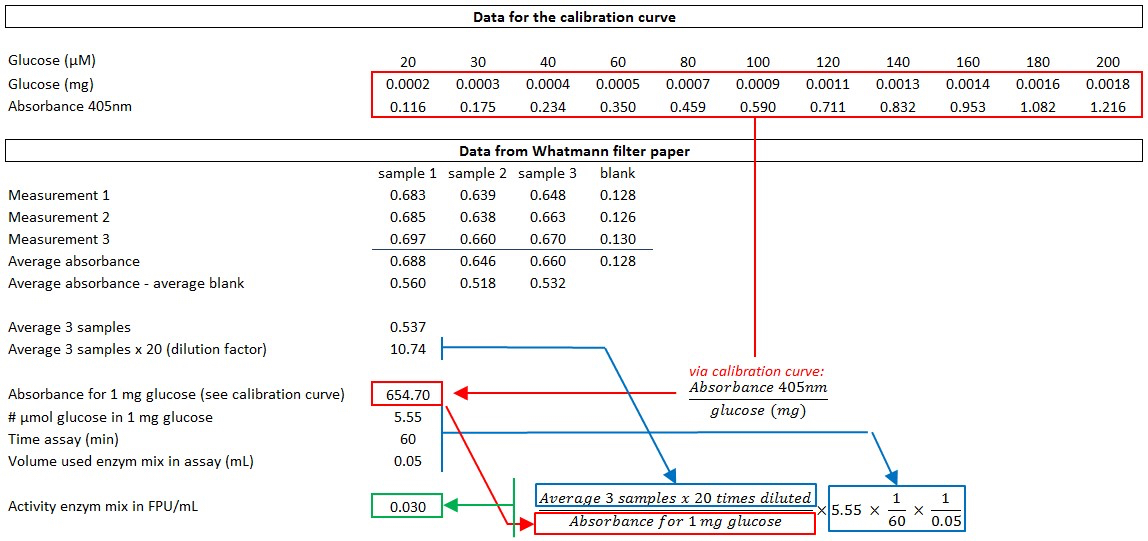
Figure 3. An example of the calculation of the enzyme activity
- Fill three safe-lock tubes with 4.5 mg of Whatman filter paper. Also include a blank, i.e. a safe-lock tube with no filter paper.
- Acid pretreatment (see Note 1)
- Weigh 10 mg aliquots of biomass. The biomass can be chopped into little pieces, e.g. Arabidopsis stem pieces of 2 mm long, or the biomass can be grinded, e.g. poplar stems.
- Add 1 ml of 1 M HCl and incubate at 80 °C for 2 h while shaking at 12.5 Hz.
- Centrifuge for 5 min at 23,477 x g and remove the supernatant.
- Wash the pellet three times by adding each time 1 ml of milliQ-water, centrifuging for 5 min at 23,477 x g and removing the supernatant.
Note 1: There are many different pretreatments possible. Broad categories are alkali or acid or hot water. Pretreatment can also be skipped. The best type of pretreatment is case by case dependent. An alkali pretreatment has the characteristic to breakdown part of lignin and ester bonds, whereas acid pretreatments will mainly breakdown glycosidic bonds in hemicellulose and amorphous cellulose.
- Weigh 10 mg aliquots of biomass. The biomass can be chopped into little pieces, e.g. Arabidopsis stem pieces of 2 mm long, or the biomass can be grinded, e.g. poplar stems.
- Alkali pretreatment
- Weigh 10 mg aliquots of biomass. The biomass can be chopped into little pieces, e.g. Arabidopsis stem pieces of 2 mm long, or the biomass can be grinded, e.g. poplar stems.
- Add 1 ml of 62.5 mM NaOH, as described in Van Acker et al. (2014) and incubate at 90 °C for 3 h while shaking at 12.5 Hz.
- Centrifuge for 5 min at 23,477 x g and remove the supernatant.
- Wash the pellet three times by adding each time 1 ml of milliQ-water, centrifuging for 5 min at 23,477 x g and removing the supernatant.
- Weigh 10 mg aliquots of biomass. The biomass can be chopped into little pieces, e.g. Arabidopsis stem pieces of 2 mm long, or the biomass can be grinded, e.g. poplar stems.
- Severe washing step
This washing step removes residual chemicals used during pretreatment, but also free sugars and other solutes that are not part of the cell wall. This step should thus also be included for samples that will be saccharified without any pretreatment.- Add 1 ml of 70% ethanol and incubate at 55 °C overnight.
- Centrifuge for 5 min at 23,477 x g and remove the supernatant.
- Wash the pellet three times by adding each time 1 ml of 70% ethanol, centrifuging for 5 min at 23,477 x g and removing the supernatant.
- Finally wash the pellet with 1 ml acetone, centrifuge for 5 min at 23,477 x g and remove the supernatant.
- Dry the pellet using a vacuum concentrator.
- Weigh the left-over biomass.
- Add 1 ml of 70% ethanol and incubate at 55 °C overnight.
- Saccharification (see Note 2)
- Dissolve every dried (pretreated or not) sample in 1 ml acetic acid buffer solution (pH 4.8) and incubate at 50 °C while shaking (12.5 Hz).
- After 5 min of incubation, add 100 µl of enzyme mix to each sample.
- At several time points, e.g. 2 h, 6 h, 24 h, 48 h after adding the enzyme mixture, take a 20 µl aliquot of the supernatant after spinning down the samples in a benchtop microcentrifuge.
- Dilute this 20 µl-aliquot in an appropriate way (i.e., in a range of 10 till 40-fold dilution) with acetic acid buffer solution (pH 4.8) and boil the diluted solution for 5 min at 99 °C to stop the enzymatic reactions.
- Measure the glucose concentration according to step H.
Note 2: To work very precisely, it is recommended to process only 2 samples every minute. This means adding enzyme to the first two samples in the first minute, adding enzyme to the next two samples in the second minute, etc. After exactly incubating for a certain time, e.g. 2 h, the first two samples are centrifuged and an aliquot is taken in the first minute. Samples are boiled to stop the reaction. In the second minute, the two next samples are centrifuged, an aliquot is taken and boiled to stop the reaction, etc.
- Dissolve every dried (pretreated or not) sample in 1 ml acetic acid buffer solution (pH 4.8) and incubate at 50 °C while shaking (12.5 Hz).
- Measuring released glucose
- For each sample (resulting from step G), standard glucose concentration (see step B) or enzyme activity measurement (see step C), transfer 50 µl to a separate well of a 96-well flat bottom microplate. All measurements are performed in triplicate.
- Fill at least 3 wells with blanks, i.e. 50 µl acetic acid buffer (pH 4.8). Also add to these blanks 150 µl GOD-POD-solution.
- Add to each well 150 µl GOD-POD-solution.
- Close the microplate with a sealing film and incubate the microtiter plate in an oven at 37 °C for exactly 30 min.
- Gently remove the sealing film and read the absorbance at 405 nm using a plate spectrophotometer.
- Use the GOD-POD calibration curve (see step B) to calculate the glucose concentration in the samples (see Notes 3, 4 and 5).
Note 3: For the calculations, take also into account the dilution factor and the starting volumes at the different time points.
Note 4: Figure 4 shows an example of saccharification results.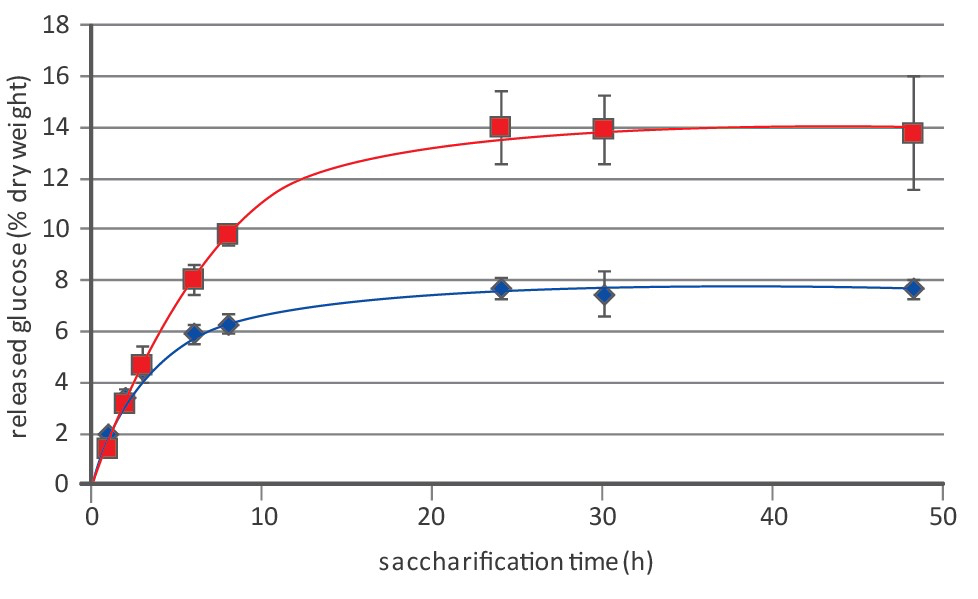
Figure 4. An example of saccharification results of WT (blue diamonds and lines) and lignin-deficient Arabidopsis thaliana (red squares and lines)
Note 5: It is common to normalize the released glucose to the theoretical amount of glucose that would be recovered if cellulose is completely saccharified. As such, the cellulose-to-glucose conversion can be calculated. To determine the cellulose content in small samples, the protocol as described by Foster and colleagues4 can be followed. An anhydroglucose unit of cellulose (162.14 g/mol) gives rise to glucose (180.155 g/mol) upon saccharification. Therefore, the theoretical complete conversion of 1 mg cellulose will give rise to 1.11 mg of glucose.
- For each sample (resulting from step G), standard glucose concentration (see step B) or enzyme activity measurement (see step C), transfer 50 µl to a separate well of a 96-well flat bottom microplate. All measurements are performed in triplicate.
Recipes
- 0.1 M acetic acid buffer solution (pH 4.5)
Prepare the acetic acid buffer pH 4.5 as follows:
32.5 ml 1 M glacial acetic acid
2.38 g sodium acetate trihydrate
967.5 ml milliQ-water - 0.1 M acetic acid buffer solution (pH 4.8)
Prepare the acetic acid buffer pH 4.8 as follows:
24.1 ml 1 M glacial acetic acid
3.52 g sodium acetate trihydrate
975.9 ml milliQ-water - GOD-POD solution (see Note 6)
Prepare the GOD-POD solution as follows:
50 mg (=1 tablet) ABTS
44.81 mg glucose oxidase
173 µl 4% peroxidase (see Note 7)
Add to reach a total volume of 100 ml with 0.1 M acetic acid buffer (pH 4.5)
Note 6: First dissolve one ABTS tablet in a volumetric flask of 100 ml in approximately 40 ml of 0.1M acetic acid buffer (pH 4.5). Once it is dissolved and glucose oxidase and peroxidase are added, adjust the volume to the mark of 100 ml with 0.1 M acetic acid buffer (pH 4.5).
Note 7: 4% peroxidase = 20 mg peroxidase dissolved in 500 µl of 0.1 M acetic acid buffer (pH 4.5).
Acknowledgments
This work was supported by grants from the Multidisciplinary Research Partnership “Biotechnology for a sustainable economy” of Ghent University, the European Commission through the Directorate General Research within the 7th Framework Program RENEWALL (KBBE-2007-3-1-01) and MultiBioPro (grant agreement N° 311804) and the Agency for Innovation by Science and Technology (IWT). RV is indebted to the Research Foundation-Flanders (FWO) for a postdoctoral fellowship.
References
- Bergmeyer, H. U. (1974). Methods of enzymatic analysis. Academic Press.
- Foster, C. E., Martin, T. M. and Pauly, M. (2010). Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J Vis Exp (37).
- Van Acker, R., Leple, J. C., Aerts, D., Storme, V., Goeminne, G., Ivens, B., Legee, F., Lapierre, C., Piens, K., Van Montagu, M. C., Santoro, N., Foster, C. E., Ralph, J., Soetaert, W., Pilate, G. and Boerjan, W. (2014). Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc Natl Acad Sci U S A 111(2): 845-850.
- Van Acker, R., Vanholme, R., Storme, V., Mortimer, J. C., Dupree, P. and Boerjan, W. (2013). Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6(1): 46.
- Xiao, Z., Storms, R. and Tsang, A. (2004). Microplate-based filter paper assay to measure total cellulase activity. Biotechnol Bioeng 88(7): 832-837.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Acker, R. V., Vanholme, R., Piens, K. and Boerjan, W. (2016). Saccharification Protocol for Small-scale Lignocellulosic Biomass Samples to Test Processing of Cellulose into Glucose. Bio-protocol 6(1): e1701. DOI: 10.21769/BioProtoc.1701.
Category
Plant Science > Plant biochemistry > Carbohydrate
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










