- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protocol-In vitro T Cell Proliferation and Treg Suppression Assay with Celltrace Violet
Published: Vol 6, Iss 1, Jan 5, 2016 DOI: 10.21769/BioProtoc.1698 Views: 33989
Reviewed by: Ivan ZanoniAndrea PuharAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Functional Phenotyping of Lung Mouse CD4+ T Cells Using Multiparametric Flow Cytometry Analysis
Céline M. Maquet [...] Bénédicte D. Machiels
Sep 20, 2023 4206 Views

T-Cell-Based Platform for Functional Screening of T-Cell Receptors Identified in Single-Cell RNA Sequencing Data Sets of Tumor-Infiltrating T-Cells
Aaron Rodriguez Ehrenfried [...] Rienk Offringa
Apr 20, 2024 6409 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3551 Views
Abstract
Measurement of the incorporation of radionuclides such as 3H-thymidine is a classical immunological technique for assaying T cell proliferation. However, such an approach has drawbacks beyond the inconvenience of working with radioactive materials, such as the inability of bulk radionuclide incorporation measurements to accurately quantitate T cell divisions, and an inability to combine proliferation analyses with simultaneous evaluation of the expression of cellular markers in divided cells. By labeling T cells with reactive dyes such as CFSE, Celltrace Violet, and others that are partitioned equally between daughter cells at each cell division, one can relatively easily track generations of proliferated cells and their expression of various molecules by flow cytometry.
FoxP3+ regulatory T cells (Treg) are critical mediators of immune tolerance and evaluation of their functionality is an important step in characterizing many immune models (Rudensky, 2011). Classically CD4+ Treg and conventional or “responder” T cells have been isolated based on their surface expression of CD25 (Treg: CD4+CD25+, Tresp: CD4+CD25-). However, we and others have noted that populations of CD4+CD25- cells express the FoxP3 transcription factor and have suppressive function. Therefore we have utilized the transgenic FoxP3-EGFP mouse to facilitate live purification of suppressor and responder populations based on EGFP (and thus FoxP3) expression. Here we present our adapted protocol for assaying regulatory T cell suppression of Celltrace Violet-labeled responder T cells.
Materials and Reagents
- 70 μm nylon mesh cell strainers (Thermo Fisher Scientific, FisherbrandTM, catalog number: 22363548 )
- 5 ml polypropylene round-bottom FACS tubes (BD Falcon®, catalog number: 352063 )
Note: Currently, it is “Corning, catalog number: 352063”. - 50 m conical polypropylene tubes (BD Falcon®, catalog number: 352098 )
Note: Currently, it is “Corning, catalog number: 352098”. - 96 well plate, U bottom (BD, Falcon®, catalog number: 353077 )
Note: Currently, it is “Corning, catalog number: 353077”. - Mice
- Biosure preservative-free 8x sheath fluid concentrate (for cell sorter) [Cedarlane, catalog number: 1027(BS) ]
- Hank’s Balanced Salt solution (HBSS) (Life Technologies, InvitrogenTM, catalog number: 14170 )
Note: Currently, it is “Thermo Fisher Scientific, catalog number: 14170”. - Fetal bovine serum (Life Technologies, InvitrogenTM, catalog number: 16170078 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 16170078”. - 1 M HEPES (Life Technologies, InvitrogenTM, catalog number: 15630080 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 15630080”. - PE anti-mouse CD8a (clone 53-6.7) (eBioscience, catalog number: 120081 )
- APC anti-mouse CD4 (clone GK1.5) (eBioscience, catalog number: 170041 )
- PerCP-eFluor 710 anti-mouse CD4 (clone RM4-5) (eBioscience, catalog number: 460042 )
- APC-eFluor 780 anti-mouse TCRβ (clone H57-597) (eBioscience, catalog number: 475961 )
- Ultracomp eBeads (eBioscience, catalog number: 012222 )
- “Fc blocking” antibody, anti-mouse CD16/CD32 (clone 2.4g2) (9.6 mg/ml) (Bio X cell, catalog number: CUS-HB-197 )
- Functional grade purified anti-mouse CD3ε (clone 145-2C11) (eBioscience, catalog number: 160031 )
- Celltrace violet cell proliferation kit (Life Technologies, InvitrogenTM, catalog number: C34557 )
Note: Currently, it is “Thermo Fisher Scientific, Molecular ProbesTM, catalog number: 16170078”. - LIVE/DEAD Fixable Yellow Dead cell stain kit (Life Technologies, InvitrogenTM, catalog number: L34959 ) or 7-Aminoactinomycin D (7-AAD) (Sigma-Aldrich, catalog number: A9400)
Note: Currently, it is “Thermo Fisher Scientific, Molecular ProbesTM, catalog number: L34959”. - 18 MΩ.cm deionized water
- Dulbecco modified Eagle medium (DMEM) (Life Technologies, InvitrogenTM, catalog number: 11965 )
Note: Currently, it is “Thermo Fisher Scientific, InvitrogenTM, catalog number: 11965 ”. - Pen/strep (100 U/ml Penicillin, 100 μg/ml Streptomycin) (Life Technologies, InvitrogenTM, catalog number: 15140122 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 15140122”. - Non-essential amino acids (Life Technologies, InvitrogenTM, catalog number: 11140050 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 11140050”. - Sodium pyruvate (Life Technologies, InvitrogenTM, catalog number: 11360070 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 11360070”. - L-glutamine (Life Technologies, InvitrogenTM, catalog number: 25030081 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 25030081”. - 2-mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- Enriched DMEM (E-DMEM) (see Recipes)
- Phosphate-buffered saline (PBS) (see Recipes)
- ACK lysis buffer (see Recipes)
Equipment
- Cell sorter (BD Biosciences, model: BD Influx )
- Flow cytometer with 405 nm excitation capability (see note 3) (BD Biosciences, model: BD LSR II )
- 37 °C humidified incubator, 5% CO2
- Centrifuge (capable of spinning 50 ml conical tubes, 5 ml FACS tubes and 96 well plates)
- Multichannel pipette (30-300 μl) (Eppendorf)
Procedure
- Cell preparation
Note: All procedures involving animals must be performed according to approved institutional, regional or national protocols (as applicable) by appropriately trained and certified personnel.- Euthanize mice by cervical dislocation and aseptically remove the spleen.
- Prepare splenocytes via standard methods. We use firm but gentle disruption of spleens through a 70 μm nylon mesh filter basket with the plunger from a 3 ml syringe, keeping tissue moist and washing cells through with HBSS + 2% FBS at room temperature. For red blood cell lysis, pellet the cells by centrifuging at ~320 x g for 5-10 min @ RT. Resuspend cell pellet in 2-5 ml ACK lysis buffer at room temperature for 5 min with periodic mixing by swirling, followed by adding ~10 volumes (20-50 ml) of HBSS + 2% FBS and spinning at ~320 x g for 5-10 min @ RT.
- Resuspend cells in HBSS + 2% FBS at room temperature. Add Fc blocking antibody and wait 5 min. Add APC anti-CD4 and PE anti-CD8 antibodies and incubate at RT for ~30 min. [Typically we resuspend splenocytes from 4 adult mice in 2 ml buffer and use 25 μl Fc block antibody stock (0.24 mg), 25 μl APC anti-CD4 (5 μg), 50 μl PE-anti CD8 (10 μg).]
- Spin cells at ~320 x g for 5-10 min @ RT. Wash cells 1x with at least 10 staining volumes (i.e.,: > 20 ml) of HBSS + 2% FBS.
- Resuspend splenocytes in sufficient HBSS + 20% FBS + 10 mM HEPES to give a concentration of ~15 x 106 cells/ml. Filter cells through a 70 μm nylon mesh filter and place cells on ice.
- Proceed to sort the CD4 single positive (SP) GFP- and CD4 SP GFP+ cell populations, directly into separate 5 ml FACS tubes containing about 1 ml of 99% FBS + 1% 1 M HEPES (final concentration 10 mM). Exclude doublets based on FSC-H vs FSC-A plots. Emphasize purity over yield. Store tubes containing sorted cells on ice after removing from the sorter. We have found it is not necessary to chill the tubes during collection on the sorter.
- Evaluate the purity of the sorted cell populations by flow cytometry. Typical yield of CD4+ GFP+ cells from one wild-type mouse spleen after sorting is approximately 7 x 105 cells. Typical yield of CD4+ GFP- cells from one wild-type mouse spleen after sorting is approximately 5 x 106 cells.
- Euthanize mice by cervical dislocation and aseptically remove the spleen.
- Assay setup
- Count, spin cells at ~320 x g for 5-10 min @ RT, and resuspend the CD4+ GFP+ (FoxP3+, “suppressors”) in E-DMEM at a concentration of (2 x 106 cells/ml). Place in a humidified 37 °C 5% CO2 incubator with the lid of the tube loose while preparing the responder cells.
- Count and wash (spin cells at ~320 x g for 5-10 min @ RT, then resuspend in the solution specified) the CD4+ GFP- (FoxP3-, “responders”) in HBSS or PBS (without serum) twice. Label approximately twice the number of cells you will need for the assay, as it is common to lose a significant proportion of your cells during the Celltrace violet (CTV) labeling process. Set aside an aliquot of cells that are unlabeled (~5 x 105 cells, resuspended in 500 μl of E-DMEM) for use as 2 controls for later flow cytometric analysis (non-CTV stained, vital stain only; and completely unstained; Figure 1 wells E7 and E8 respectively). These cells should be stored at 37 °C in a humidified 5% CO2 incubator-they can be added directly to the appropriate wells of the assay plate now or stored until assay setup in a 15 ml or 50 ml conical tube in the incubator with the cap on loosely. See Note 4 for discussion of the use of non-fractionated splenocytes as responders.
- Label the CD4+ GFP- cells with CTV. Briefly, prepare a 5 mM CTV stock solution according to the manufacturer’s directions by adding 20 μl of the supplied (room temperature) DMSO to an aliquot of dry CTV dye and mixing thoroughly. Prepare a 5 μM working solution by diluting stock 1/1,000 in prewarmed (37 °C) HBSS or PBS. Resuspend pelleted, washed CD4+GFP-cells in the working solution at a concentration of between 1-15 x 106 cells/ml, ensuring complete and rapid cell pellet disruption/ re-suspension by triturating through a serological pipette. Incubate for 20 min in a 37 °C water bath.
- Quench the labeling reaction by adding 5 volumes of cold DMEM + 10% FBS. Incubate for 5 min at RT, then wash cells 1x with E-DMEM and resuspend in prewarmed (37 °C) E-DMEM. Incubate at 37 °C in a humidified 5% CO2 incubator for at least 15 min.
- Wash cells with 25-50 ml warmed E-DMEM, count and resuspend at 1 x 106 cells/ml in E-DMEM.
- While performing the labeling reaction, prepare antigen presenting cells (APC) for the plate assay by euthanizing a Rag KO mouse, removing the spleen, and preparing splenocytes as previous with RBC lysis. Wash, count, and resuspend these cells at 2 x 106 cells/ml in E-DMEM.
- Set up assay plate (see Table 1 and Figure 1):
Each well used of the 96 well U-bottom assay plate should contain:- 1 x 105 Responder T cells
- Either 1 x 105 (Treg: Tresp ratio 1:1), 5 x 104 (1:2), 2.5 x 104 (1:4), 1.25 x 104 (1:8), 6.25 x 103 (1:16) or 0 (0:1) suppressor T cells
- 1 μg/ml anti-CD3ε antibody
- 1 x 105 Antigen presenting cells (RBC-depleted splenocytes from Rag KO animal(s), prepared in step B13 above)
- Sufficient E-DMEM media to give a final volume of 250 μl per well.
Notes: Each condition should be assayed at least in triplicate. Try to avoid using the edge wells of the assay plate as they are subject to increased evaporation rates. Remember to include 2 sets of wells containing no Treg and no anti-CD3ε stimulation: One set will serve as the unstimulated control (Figure 1, wells B6-B9), the other will be used as a compensation control for the CTV stain during later flow cytometric analysis (Figure 1, wells D7-D9). Add the non-CTV labeled cells saved from step 9 to two wells of the assay plate (Figure 1, wells E7-E8).
Table 1. Suppression assay set up guide-each well contains the following volumes of components for each condition in the left column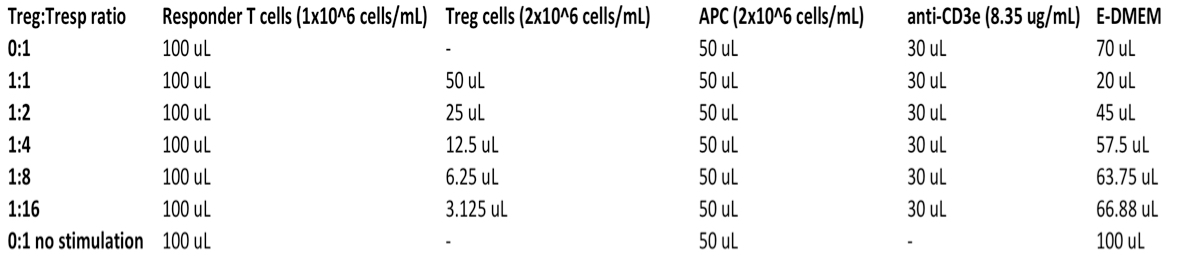
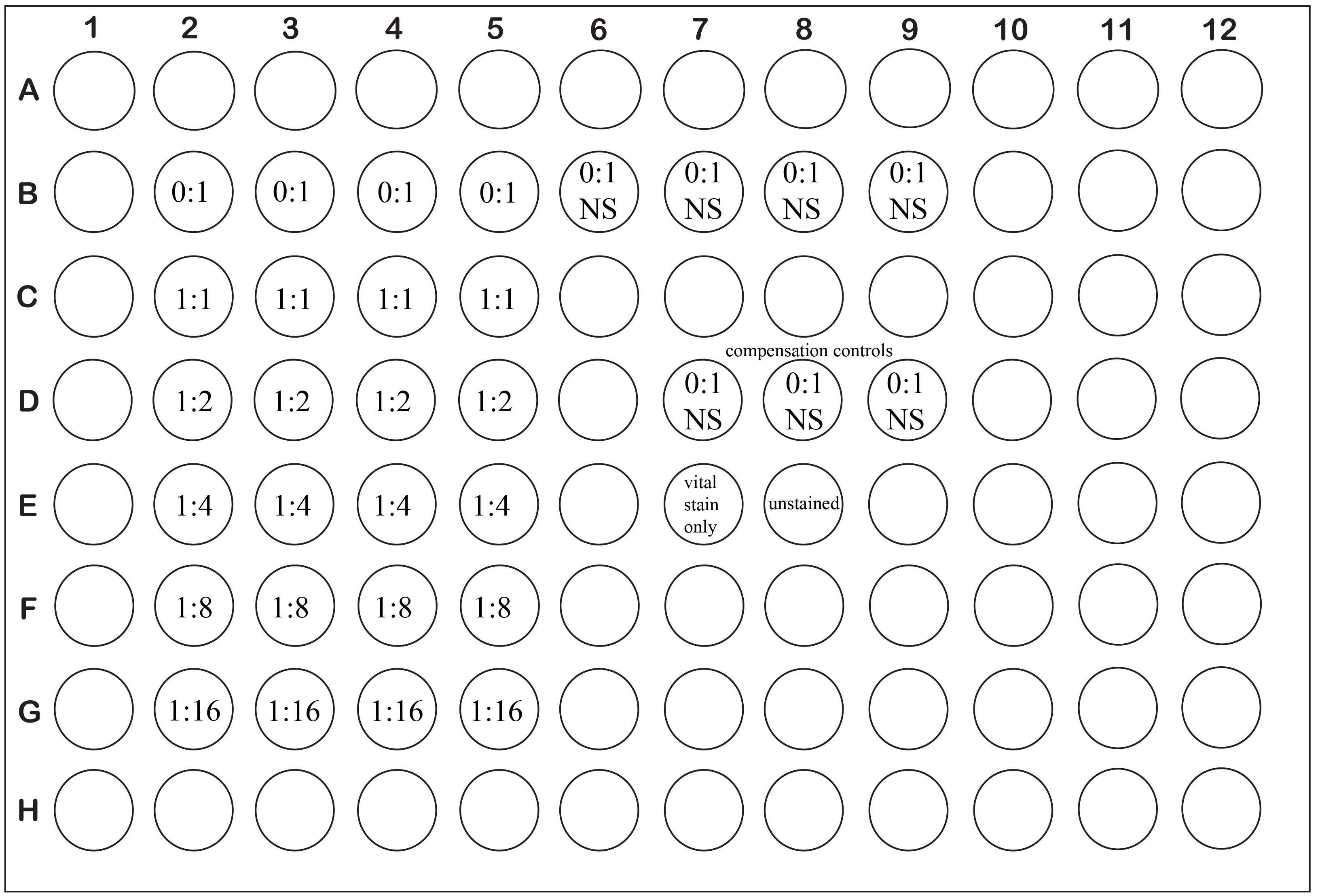
Figure 1. Example plate layout for a typical suppression assay with experimental wells set up in quadruplicate. Ratios are expressed as Treg: Tresponders.
- 1 x 105 Responder T cells
- Incubate the assay plate in a 37 °C, 5% CO2 humidified incubator for 5 d.
- Count, spin cells at ~320 x g for 5-10 min @ RT, and resuspend the CD4+ GFP+ (FoxP3+, “suppressors”) in E-DMEM at a concentration of (2 x 106 cells/ml). Place in a humidified 37 °C 5% CO2 incubator with the lid of the tube loose while preparing the responder cells.
- Staining
- Centrifuge the assay plate for 5 min at ~400 x g.
- Remove the supernatant from the wells with a quick “flicking” motion into an approved receptacle.
- Using a multichannel pipette, add 200 μl HBSS with no serum (if labeling cells with fixable yellow live/dead stain) or 200 μl HBSS + 2% FBS (if using 7-AAD for dead cell discrimination) to each well to wash. Pipet up and down ~3 times to resuspend the cells. Centrifuge the assay plate again for 5 min at ~400 x g and flick off the supernatant.
Note: If you are using fixable yellow live/dead stain in your assay perform steps 19-22, otherwise skip to step 23. - Prepare sufficient fixable yellow live/dead stain in HBSS according to the manufacturer’s protocol such that 200 μl is available per well of the plate that was used. Briefly, 50 μl of the supplied DMSO is added to one vial of the dry dye solution and mixed. This reconstituted dye solution is then diluted 1/1,000 in HBSS for use in labeling.
- Resuspend cells in each well (excluding the CTV-only control wells and unstained control wells, Figure 1 wells D7-D9 and E8) in 200 μl LIVE/DEAD fixable yellow stain/ HBSS by pipetting up and down with a multichannel pipette. Store plate on ice in the dark for 30 min.
- Centrifuge the plate for 5 min at ~400 x g, remove supernatant, and wash with 200 μl/well HBSS + 2% FBS.
- Centrifuge the plate for 5 min at ~400 x g, remove supernatant.
- Proceed with staining in the plate using your desired panel of antibodies with or without an Fc blocking step.
- Centrifuge the assay plate for 5 min at ~400 x g.
- Example staining
- Add 30 μl Fc block cocktail to each experimental (non-compensation control) well (1 μl Fc block antibody stock, 29 μl HBSS + 2% FBS, final 9.6 μg of Fc block per well). Pipet up and down to mix. Incubate at RT for 5 min. For larger experiments it is handy to do this with a multichannel pipette by preparing the cocktail and distributing it across several wells of a source 96 well plate.
- Without washing, add 30 μl surface marker staining cocktail per experimental well and pipet up and down to mix. Incubate at room temperature for ~30 min.
Example surface staining cocktail (per staining, all Abs purchased from eBioscience):
1 μl PE anti-mouse CD8a (0.2 μg)
0.25 μl PerCP-eFluor 710 anti-mouse CD4 (0.05 μg)
0.75 μl APC-eFluor 780 anti-mouse TCRβ (0.15 μg)
28 μl HBSS + 2% FBS - While the cells are incubating, create single-stained compensation controls for each antibody used in your flow cytometry experiment using Ultracomp eBeads in 5 ml round bottom FACS tubes according to the manufacturer’s protocols.
- Wash wells 2x with HBSS + 2% FBS. Resuspend in 200 μl per well HBSS + 2% FBS for analysis. If using 7-AAD for dead cell discrimination, resuspend all wells except for the CTV control and unstained control wells in 200 μl per well HBSS + 2% FBS + 1 μg/ml 7-AAD. (Resuspend the CTV control and unstained control wells in 200 μl HBSS + 2% FBS with no 7-AAD.)
- Analyze on flow cytometer. For each well we typically remove the 200 μl per well from the plate, transfer it to a 5 ml round-bottom FACS tube containing 400 μl HBSS + 2% FBS, and load on the cytometer for acquisition. We generally analyze cells without fixation, however one could fix cells with ~1% formaldehyde in PBS prior to analysis if desired or required by the local flow cytometry facility. Please note that fixation is not compatible with using 7-AAD for dead cell discrimination, and will generally destroy GFP fluorescence in GFP+ cells.
- Add 30 μl Fc block cocktail to each experimental (non-compensation control) well (1 μl Fc block antibody stock, 29 μl HBSS + 2% FBS, final 9.6 μg of Fc block per well). Pipet up and down to mix. Incubate at RT for 5 min. For larger experiments it is handy to do this with a multichannel pipette by preparing the cocktail and distributing it across several wells of a source 96 well plate.
- Analysis tips and sample data
We suggest drawing a gate containing the lymphocytes based on forward and side scatter (FSC/SSC), and if dead cell discrimination is desired, gating the live cells (i.e.,: LIVE/DEAD fixable yellow stain negative/low population, or 7-AAD low population, see Figures 2A, B respectively). The unstimulated wells will have a clear population of dead cells to help draw the dead cell gate. Then gate the TCRβ+, CD4 SP cells. We have noted that using anti-CD3ε (clone 145-2C11) for stimulation decreases staining intensity for TCRβ (using the H57-597 clone), keep this in mind when drawing gates for TCRβ+, CD4 SP cells in the stimulated and unstimulated conditions.
Next, in order to analyze the extent of proliferation, examine this gated TCRβ+, CD4 SP population for CTV (Figure 2). Wells that contain Treg (which are not labeled with CTV) will have a population of TCRβ+CD4+ cells that were not stained with CTV and must be excluded from the analysis-use the unstained sample along with the 1:1 Treg:Tresp samples (which should show low responder proliferation allowing easy delineation of Treg vs responder populations) to determine the threshold to place a gate encompassing all CTV-positive events (see Figure 2C for clarification). One cannot exclude Treg on the basis of FoxP3 positivity as some Treg may have lost FoxP3 expression during the assay. Then use the unstimulated samples (i.e.,: Figure 1, wells B6-B9) to draw a gate encompassing all CTV-positive events that have proliferated at least once (Figure 2D). If the assay has incubated for too long, some extensively proliferated cells may have lost enough dye to become indistinguishable from CTV-negative cells. In this case we suggest shortening your assay incubation time to avoid this issue. We are able to resolve 7 generations of cell division and thus we report the data as the percent of generation 0 to 7 cells that have proliferated at least once. It is also important to present the proliferation data visually, as in Figure 2E.
Figure 2. Analysis of Celltrace Violet staining following suppression assay incubation and sample data. A. Example of gating live cells for analysis based on negative/low staining with LIVE/DEAD yellow fixable stain or B. 7-AAD. C. Examining a sample containing a high ratio of Treg to responder T cells facilitates placement of a gate encompassing true CTV labeled responders and avoiding the unlabeled Treg in the assay. D. Example of gating of CTV+ cells from generations 0-7 that have proliferated at least once. E. Visual comparison of the extent of responder T cell proliferation using various ratios of Treg to responder T cells.
Notes
- The FoxP3-EGFP strain used for these studies (B6.Cg-Foxp3tm2Tch/J) has been engineered to produce a bicistronic transcript from the endogenous foxp3 locus encoding the normal endogenous FoxP3 protein as well as EGFP. Another strain referred to by the same common “FoxP3-EGFP” name instead encodes a FoxP3-EGFP fusion protein at the endogenous foxp3 locus. This fusion protein has been shown to have altered function and we thus cannot recommend its use. The fusion protein strain contains the Foxp3
allele, developed by the Rudensky group, and was previously available through The Jackson Laboratory although it has since been removed from their catalog. Please see Chatila and Williams (2012) and references therein for a more comprehensive overview of the issue. - While we do not use this approach, the assay should be easily amenable to the use of Treg and responders purified based on the classical CD4+CD25+/CD4+CD25- surface expression phenotype.
- The flow cytometer used in our facility is configured to detect Celltrace violet with a 450 nm/50 nm bandpass filter set, and LIVE/DEAD fixable yellow stain with a 475 low-pass filter + 525 nm/50 nm bandpass filter set. One could substitute other vital stains instead of the LIVE/DEAD fixable yellow stain or 7-AAD as required to be detectable with one’s particular cytometer configuration/requirement for fixation.
- Instead of using CD4+FoxP3- cells as responders, one can also substitute Celltrace-violet labeled, unfractionated splenocytes as responder cells in this assay. For this approach we use 2 x 105 labeled splenocytes per well as responders, and omit adding Rag KO splenocytes as APC.
Recipes
- Enriched DMEM (E-DMEM)
10% fetal bovine serum
1% Pen/strep (100 U/ml Penicillin, 100 μg/ml Streptomycin)
1% non essential amino acids
1% sodium pyruvate
2 mM L-glutamine
50 μM 2-mercaptoethanol - Phosphate-buffered saline (PBS)
137 mM NaCl
2.7 mM KCl
10mM Na2HPO4
1.8 mM KH2PO4 - ACK lysis buffer
15.5 mM NH4Cl
1 mM KHCO3
0.01 mM EDTA
Acknowledgments
The authors would like to thank Perveen Anwar for assistance with animal care. All animal work described herein was undertaken in accordance with approved animal care protocols at the University of Alberta. This work was supported by a grant from CIHR (FRN79521) to CCA and by doctoral studentships from the Alberta Diabetes Institute and Alberta Innovates Health Solutions (AIHS) to KKE and a senior scholar award from AIHS to CCA. This work was also supported by an Alberta endMS Regional Research and Training Centre Graduate Student Support Grant of the endMS Research and Training Network from the Multiple Sclerosis Society of Canada and the Multiple Sclerosis Scientific Research Foundation.
References
- Chatila, T. A. and Williams, C. B. (2012). Foxp3: shades of tolerance. Immunity 36(5): 693-694.
- Ellestad, K. K., Thangavelu, G., Ewen, C. L., Boon, L. and Anderson, C. C. (2014). PD-1 is not required for natural or peripherally induced regulatory T cells: Severe autoimmunity despite normal production of regulatory T cells. Eur J Immunol 44(12): 3560-3572.
- Kruisbeek, A. M., Shevach, E. and Thornton, A. M. (2004). Proliferative assays for T cell function. Curr Protoc Immunol Chapter 3: Unit 3 12.
- Rudensky, A. Y. (2011). Regulatory T cells and Foxp3. Immunol Rev 241(1): 260-268.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ellestad, K. K. and Anderson, C. C. (2016). Protocol-In vitro T Cell Proliferation and Treg Suppression Assay with Celltrace Violet. Bio-protocol 6(1): e1698. DOI: 10.21769/BioProtoc.1698.
Category
Immunology > Immune cell function > Lymphocyte
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









