- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Neurite Outgrowth Assay
Published: Vol 6, Iss 1, Jan 5, 2016 DOI: 10.21769/BioProtoc.1694 Views: 12036
Reviewed by: Soyun KimYatang LiEmmanuelle Berret

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Microfluidic Cultures of Basal Forebrain Cholinergic Neurons for Assessing Retrograde Cell Death by Live Imaging
Srestha Dasgupta [...] Wilma J. Friedman
Jan 5, 2025 1861 Views

Time-Lapse Super-Resolution Imaging and Optical Manipulation of Growth Cones in Elongating Axons and Migrating Neurons
Masato Sawada [...] Kazunobu Sawamoto
Mar 20, 2025 2141 Views

Cryopreservation of Bulk-Produced Primary Rat Oligodendrocyte Progenitor Cells
Hanki Kim [...] Jun Young Choi
Jun 20, 2025 1427 Views
Abstract
Neurite outgrowth in culture provides an easy way to determine the effects of a particular substrate or exogenous factor on neuron behavior. Dissociated neurons can be plated on a variety of substrates and the length of the longest neurite outgrowth can be compared. Here, we describe how to isolate and dissociate dorsal root ganglion (DRG) neurons, culture them on coverslips, and measure longest neurite outgrowth.
Keywords: Neurite outgrowthMaterials and Reagents
- 12 mm glass coverslips
- 24 well plate
- Pipette tips
- Poly-L-lysine hydrobromide (PLL) (Sigma-Aldrich, catalog number: P1274 )
- Laminin Mouse Protein, Natural (Invitrogen, catalog number: 23017-015 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 23017-015”. - HBSS-CMF (HBSS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14185-052 )
- Collagenase II (Worthington Biochemical Corporation, catalog number: 4176 )
- Dispase II (Roche Diagnostics, catalog number: 04942078001 )
- Penicillin-streptomycin (Invitrogen, catalog number: 15140122 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 15140122”. - B-27® Serum-Free Supplement (50x) (Invitrogen, catalog number: 17504-044 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 17504-044”. - Glutamax (Thermo Fisher Scientific, GibcoTM, catalog number: 35050 )
- Neurobasal-A (Invitrogen, catalog number: 10888-022 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 10888-022”. - 1x Phosphate-buffer saline (PBS) (Roche Diagnostics, catalog number: 11-666-789-001 )
- 4% paraformaldehyde (PFA) (Electron Microscopy Sciences, catalog number: 15710-S )
- Monoclonal Anti-β-Tubulin III antibody produced in mouse (msIgG2b) (Sigma-Aldrich, catalog number: T8660 )
- Goat anti-mouse IgG2b 594 (Invitrogen, catalog number: A21145 )
Note: Currently, it is “Thermo Fisher Scientific, NovexTM, catalog number: A21145”.
Equipment
- 37 °C, 5% CO2 forced air incubator
- Dissecting microscope
- Surgical tools
- Pipettes
- Hemocytometer
- Microcentrifuge
Procedure
- Prepping coverslips
- Place 12 mm glass coverslips in a 24-well plate (1 coverslip per well).
- Coat the coverslips with poly-l-lysine (0.1 mg/ml; PLL) either overnight at room temperature or for 2 h at 37 °C.
- Remove PLL from the coverslips in the wells.
- Add ddH2O to each well to wash the coverslips.
- Remove ddH2O using a pipette or suction. Do not allow the coverslips to dry completely. Upon removing ddH2O from the coverslips, immediately move to the next step.
- Bath coverslips in laminin (5 μg/ml) in HBSS-CMF and incubate at 37 °C for 2 h before plating cells.
Notes:- This concentration of laminin can be varied to enhance or reduce outgrowth potential.
While dissociated adult DRG neurons grow well at this concentration, longer outgrowth can be obtained with higher laminin concentrations, whereas growing neurons on lower laminin concentrations will provide a more challenging substrate for outgrowth. - Laminin should remain on coverslips until cells are ready to plate. There is no need to wash the coverslips once the laminin is removed.
- Neurons may be plated directly on laminin-coated coverslips, or plated on a monolayer of cells to examine the effects of neurite outgrowth on a cellular substrate.
- This concentration of laminin can be varied to enhance or reduce outgrowth potential.
- Plating a monolayer of cells
- A monolayer of cells can be created by densely plating a cell type of interest (astrocytes, stem cells, etc.) onto the coverslip after the laminin-coating has occurred.
- To create a monolayer, concentrate 60,000 cells/60 μl of media. (This concentration can be greater or less depending on the rate of cell division for a particular cellular population.)
- Remove laminin from the coverslips (step A6).
- Place the entire 60 μl drop of cells into the center of the coverslip and allow the plate to remain untouched at RT for 30 min. This will allow the cells some time to adhere to the coverslip.
- After the 30 min incubation, gently add an additional 1 ml of media down the side of the well so to minimize the dispersal or disruption of the cell solution in the center of the slip.
- Allow the cells to incubate overnight at 37 °C before adding DRG neurons.
- A monolayer of cells can be created by densely plating a cell type of interest (astrocytes, stem cells, etc.) onto the coverslip after the laminin-coating has occurred.
- Place 12 mm glass coverslips in a 24-well plate (1 coverslip per well).
- Dissociating DRG neurons
- Remove all DRGs from an adult mouse or rat.
- Trim the central and peripheral roots.
- Place DRGs into a 1.5 ml microcentrifuge tube containing collagenase II (200 U/ml) and dispase II (2.5 U/ml) in HBSS-CMF. Dispase/collagenase helps to digest the dura surrounding the ganglia, allowing the cell bodies to fully dissociate from one another.
- Incubate for 1 h at 37 °C.
- Remove DRGs from the incubator and centrifuge the sample for 2 min, 5,600 RPM.
- Gently remove supernatant and discard. Be careful not to disturb the pellet containing the cells.
- Add fresh CMF solution (~1 ml) to the microcentrifuge tube containing the pellet, gently triturate, then centrifuge for 2 min, 5,600 RPM.
- Repeat steps B6-7 two more times (3 total washes).
- After the third wash, resuspend the dissociated neurons in 1 ml of Neurobasal-A media supplemented with B-27, GlutaMAX, and penicillin-streptomycin.
Note: Once the cells have been dissociated (step B3), it is important to complete the subsequent steps in a timely fashion (it should take approximately 20 min) to minimize neuronal death during the washes. If necessary, keep the cells on ice during this process. - Using a hemocytometer, count the number of neuronal cell bodies present in the sample.
- Plate neurons on coverslips at a concentration of 1,500-2,000 neurons per coverslip (in 1 ml Neurobasal-A per coverslip).
Note: Neurons can be plated on laminin-coated coverslips 2 h after coating them. Typically adding laminin just before beginning the DRG isolation and dissociation will allow plenty of time for the substrate to adhere to the coverslip. - Incubate at 37 °C for 24 h.
- Remove all DRGs from an adult mouse or rat.
- Plating cells for neurite outgrowth
- 24 h after DRG plating, remove the media from the wells.
- Add ~500 ml 4% PFA for 30 min at room temperature.
- Remove PFA.
- Wash 3 x 10 min with 1x PBS.
- Block coverslips overnight at 4 °C.
- Stain cultures with β-III tubulin (1:500 in block + triton) overnight at 4 °C or 2 h at room temperature.
- Wash cultures 3 times for 10 min each with 1x PBS.
- Stain with the appropriate secondary antibody overnight at 4 °C or 2 h at room temperature.
- Mount coverslips onto slides.
- To mount coverslips, use a pair of size 3 jeweler’s forceps to carefully lift the coverslip up the side of the well. Be sure to keep track of which side of the coverslip contains the cells.
- Carefully grab the edge of the coverslip with the forceps. Be careful not to grab the center of the coverslip or you will scrape off the cells.
- Flip the coverslip to be side down and set the coverslip onto the slide.
- Seal the edge of the coverslip with clear nail polish and allow the slides to dry.
- To mount coverslips, use a pair of size 3 jeweler’s forceps to carefully lift the coverslip up the side of the well. Be sure to keep track of which side of the coverslip contains the cells.
- Image all neurons with processes.
- 24 h after DRG plating, remove the media from the wells.
- Measuring neurite outgrowth
- Using MetaMorph software, measure the longest neurite per neuron. This can be done using either the traced line or multi-line tool.
- Open MetaMorph software.
Figure 1. Open screen - To measure a neurite, open “Region Measurements.”
Figure 2. Region measurements cropped - The “Region Measurement” window should open. Select “active region” to see the length of the most recent neurite measured.
Figure 3. Active region - Go to “Measure.” Click “Calibrate Distances.”
Figure 4. Calibrate distance
Note: Distance may need to be calibrated based on the scope and magnification used to image the neurons. A calibration must be established for each objective used. These calibrations will vary based on scope. Therefore, for every scope and every objective, a calibration must be determined and added to the list.
Select the calibration that corresponds to the appropriate microscope and objective used to image the neuron being measured.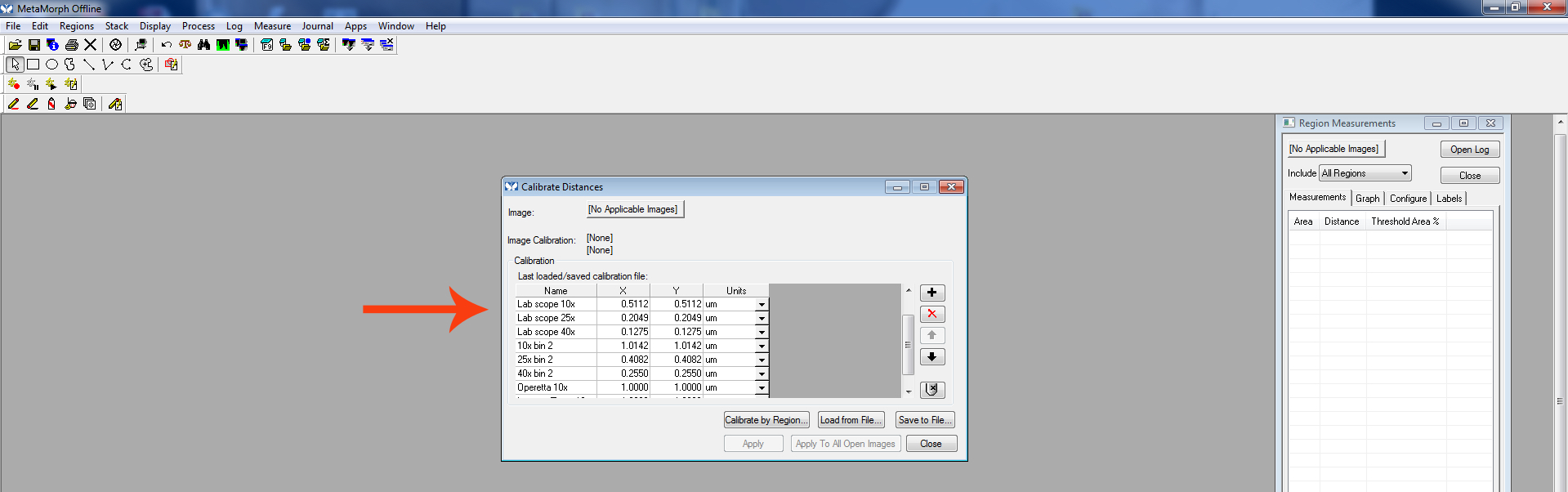
Figure 5. Calibrate distances screen - Open the image to be measured.
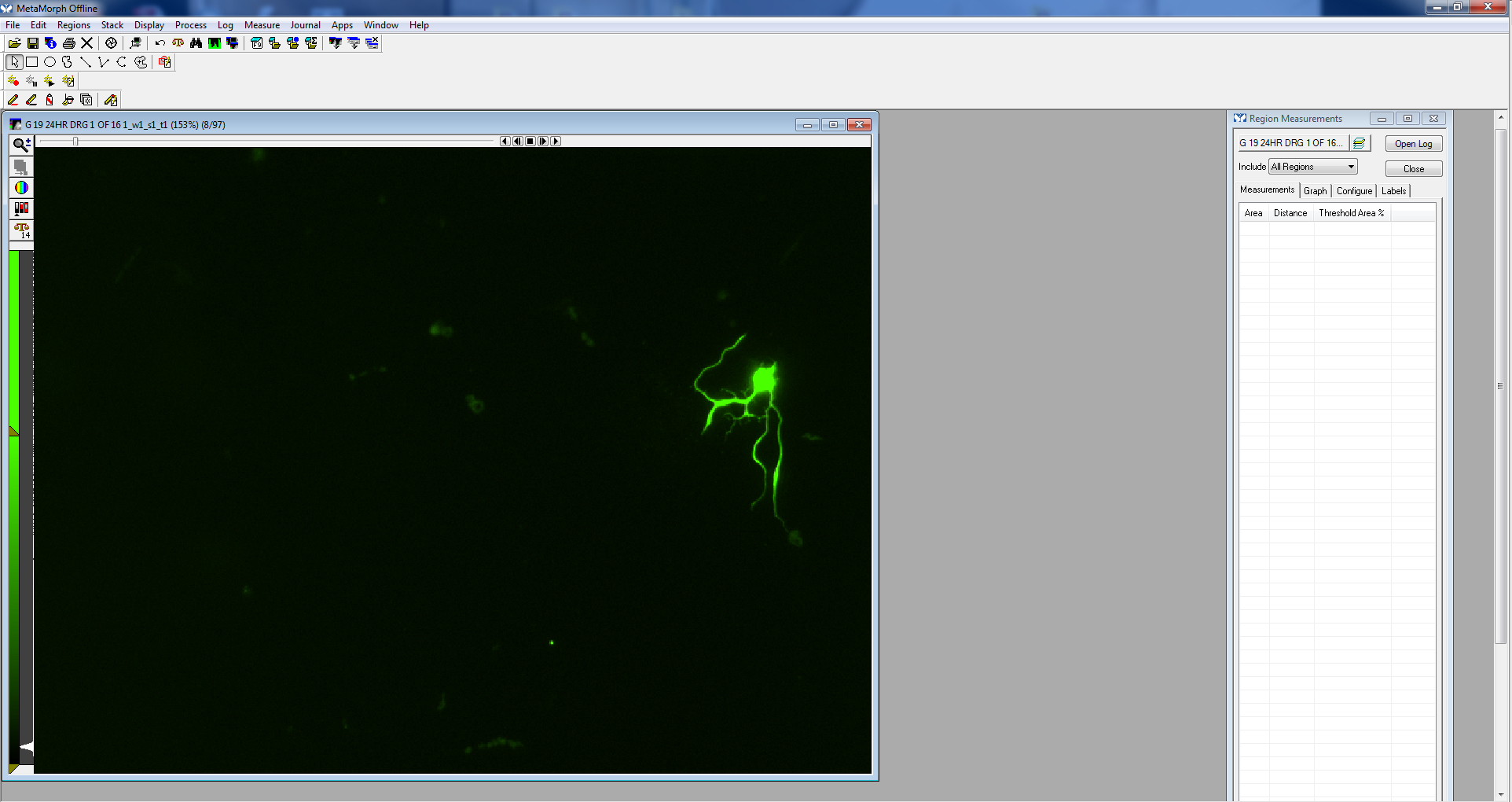
Figure 6. Image opened - Using either the traced or multi-line tool, trace a process, beginning near the cell body, extending to the tip of the process. Processes often branch, so be sure to follow the appropriate path (the longest process).
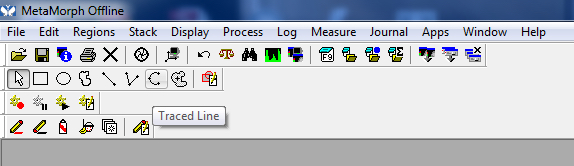
Figure 7. Traced tool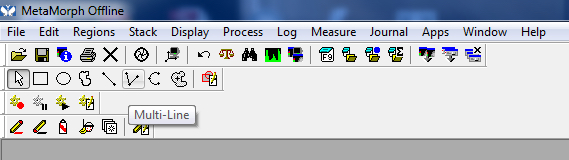
Figure 8. Multi-line tool - Once the entire process is traced, double click. This will display the “region measurement” or the length of the neurite that has just been traced. It will be displayed both on the image, as well as on the region measurements window.
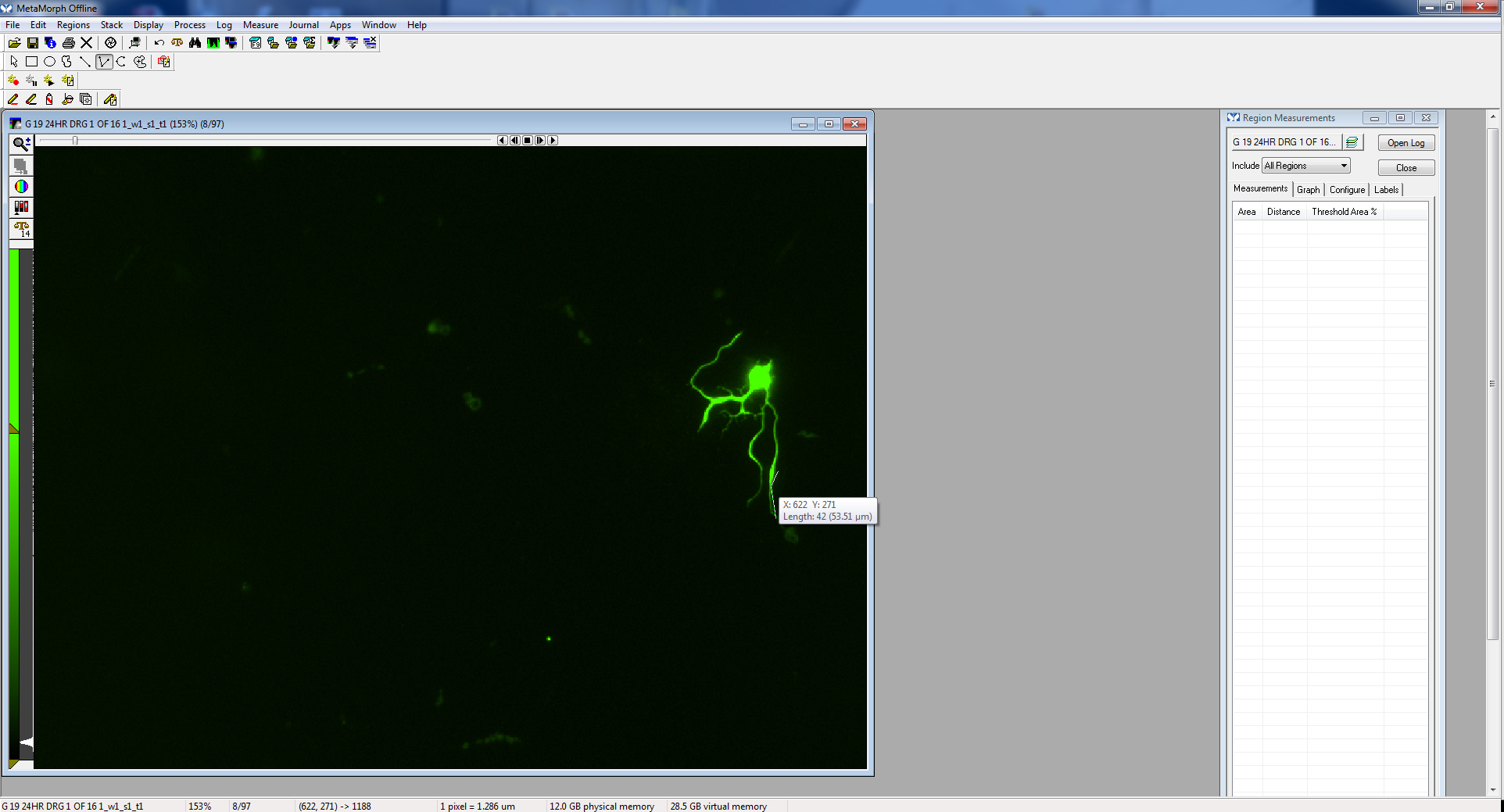
Figure 9. Begin clicking
Be sure “Active Region Measurements” is displayed in the “Region Measurements” window to see the measurement of the most recently measured neurite.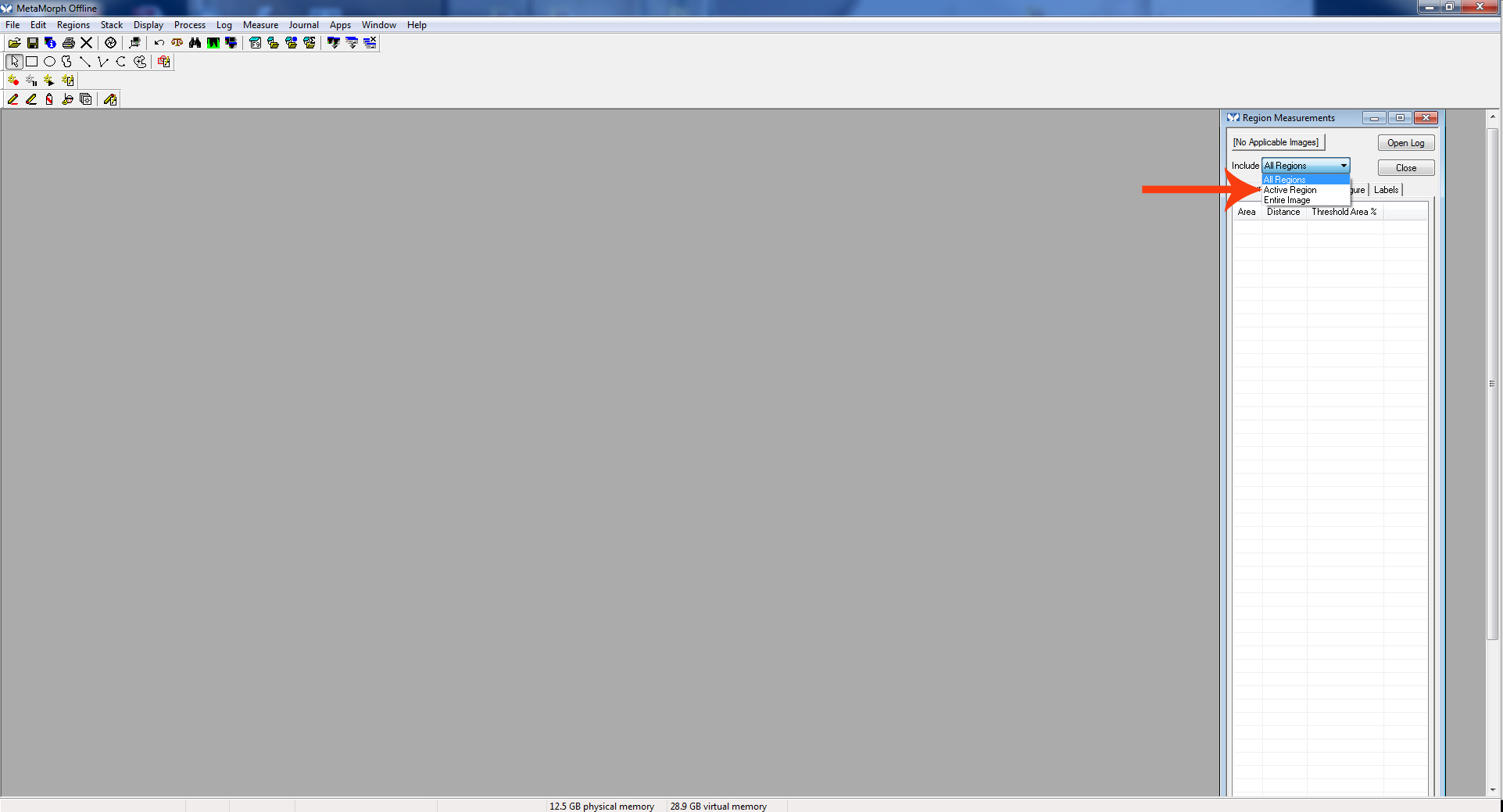
Figure 10. Active region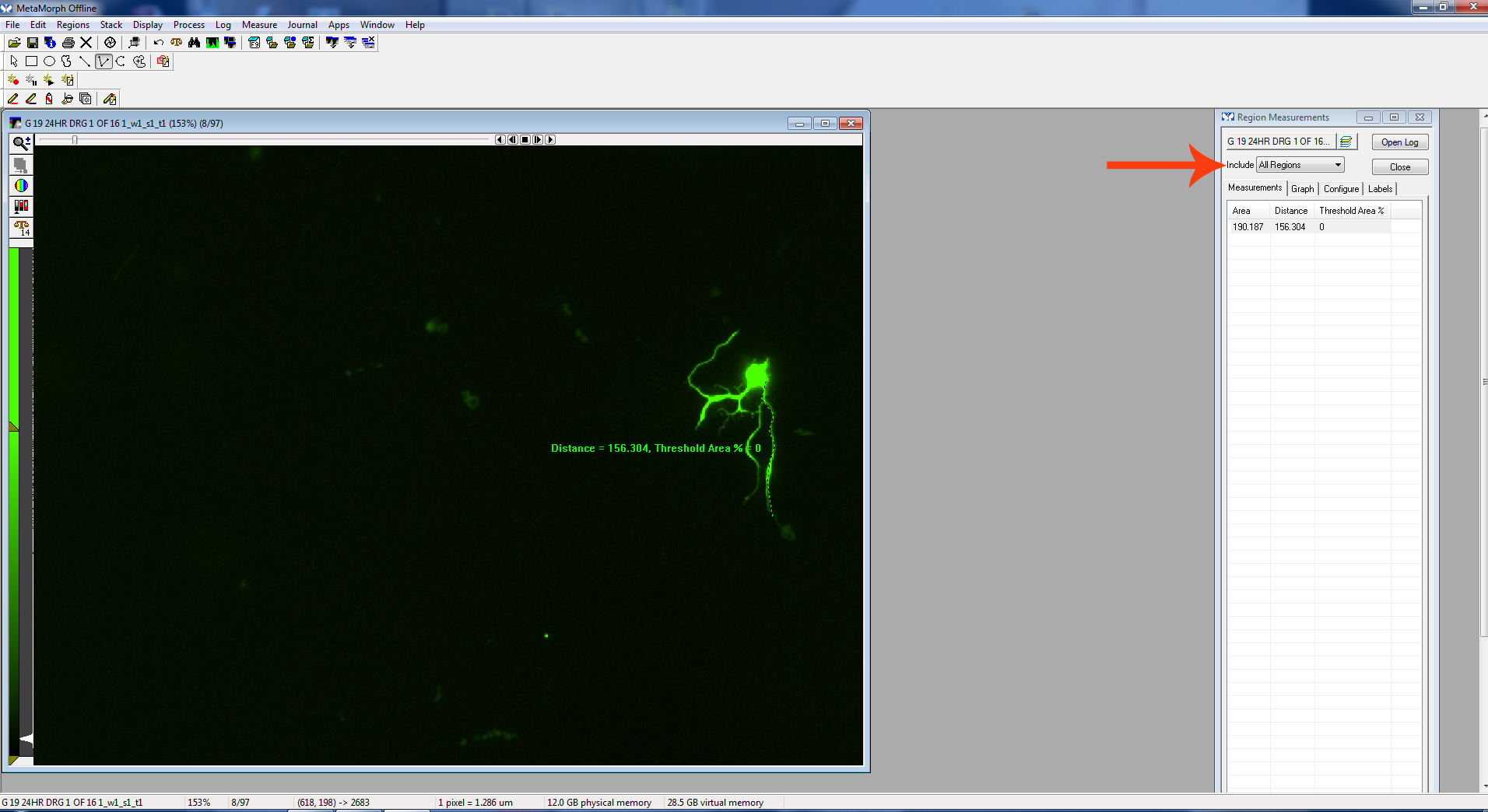
Figure 11. Finish measurement
- Open MetaMorph software.
- Using MetaMorph software, measure the longest neurite per neuron. This can be done using either the traced line or multi-line tool.
Notes
To determine the longest neurite per neuron, often multiple neurites per neuron will have to be measured, then select the longest of those measured. Depending on the density and placement of neurons, it may be difficult to determine whether a particular neurite is coming from one neuronal cell body or the other. If the cell body of origin cannot be determined, exclude those neurites/neurons from analysis.
Acknowledgments
This protocol was used in Filous et al. (2014). This work was supported by National Institutes of Health- National Institute of Neurological Disorders and Stroke Grant NS025713, National Institutes of Health Training Grant T32, and the New York State Department of Health and Brumagin Memorial Fund.
References
- Filous, A. R., Tran, A., Howell, C. J., Busch, S. A., Evans, T. A., Stallcup, W. B., Kang, S. H., Bergles, D. E., Lee, S. I., Levine, J. M. and Silver, J. (2014). Entrapment via synaptic-like connections between NG2 proteoglycan+ cells and dystrophic axons in the lesion plays a role in regeneration failure after spinal cord injury. J Neurosci 34(49): 16369-16384.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Filous, A. R. and Silver, J. (2016). Neurite Outgrowth Assay. Bio-protocol 6(1): e1694. DOI: 10.21769/BioProtoc.1694.
- Filous, A. R., Tran, A., Howell, C. J., Busch, S. A., Evans, T. A., Stallcup, W. B., Kang, S. H., Bergles, D. E., Lee, S. I., Levine, J. M. and Silver, J. (2014). Entrapment via synaptic-like connections between NG2 proteoglycan+ cells and dystrophic axons in the lesion plays a role in regeneration failure after spinal cord injury. J Neurosci 34(49): 16369-16384.
Category
Neuroscience > Cellular mechanisms > Cell isolation and culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









