- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of Rubisco from Chlamydomonas reinhardtii
Published: Vol 5, Iss 23, Dec 5, 2015 DOI: 10.21769/BioProtoc.1673 Views: 13357
Reviewed by: Maria SinetovaAgnieszka ZienkiewiczAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2870 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1990 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
Chlamydomonas reinhardtii is a model organism for chloroplast studies. Besides other convenient features, the feasibility of chloroplast genome transformation distinguishes this unicellular alga as ideal for the manipulation of chloroplastic gene expression aiming biotechnological goals, such as improved biofuel and biomass production. Ribulose 1, 5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39, Rubisco) is the photosynthetic carbon-fixing enzyme which is considered crucial for biomass accumulation in algal cultures. Purification of wild type and site-directed mutants of Rubisco in C. reinhardtii is usually performed to study its catalytic properties and assess the carbon-fixing potential of the strains. In this protocol Rubisco is extracted through sonication of cell pellets, and purified by ammonium sulfate precipitation, sucrose gradient centrifugation (Goldwaithe and Bogorad, 1975) and anion exchange chromatography.
Keywords: Ribulose 1,5-bisphosphate carboxylase/oxygenaseMaterials and Reagents
- Vacuum driven disposable filtration system (Stericup 0.22 μm) (Merck Millipore Corporation, catalog number: P25628X1 )
- Graduated cylinder (50 ml)
- Sephadex G-25 columns (PD-10) (GE Healthcare, catalog number: 17-0851-01 )
- Glass rod
- Thick wall polycarbonate plastic tubes for ultracentrifugation (Beckman Coulter, catalog number: 355631 )
- Glass disposable micro pipet (Corning Inc., catalog number: 7099S-20 )
- Resource Q anion exchange column 6 ml (GE Healthcare, catalog number: 17-1179-01 )
- Chlamydomonas reinhardtii cells (about 8 g of wet weight collected from 2-3 liters of a 1.2 x 107 cells/ml photo-heterotrophic culture)
- Tris (2-amino, 2-hydroxymethyl, 1, 3-propanediol) (Trizma base) (Sigma-Aldrich, catalog number: T-1503 )
- Magnesium sulfate heptahydrate (MgSO4.7H2O) (vidraFOC, Panreac, catalog number: 131404-1210 )
- Magnesium chloride hexahydrate (MgCl2.6H2O) (Merck Millipore Corporation, catalog number: 1.05833 )
- Sodium chloride (NaCl) (AppliChem GmbH, Panreac, catalog number: 121659.1211 )
- Sodium bicarbonate (NaHCO3) (Merck Millipore Corporation, catalog number: 1.06329 )
- 2-mercaptoethanol (Merck Millipore Corporation, catalog number: 8.05740 )
- Sulfuric acid (H2SO4) (Sigma-Aldrich, Fluka, catalog number: 84718 )
- Hydrochloric acid (HCl) (VWR International, J.T.Baker®, catalog number: 6081 )
- Sucrose (Sigma-Aldrich, Fluka, catalog number: 84100 )
- cOmplete (Protease inhibitors tablets) (Roche Diagnostics, catalog number: 11697 498001 )
- Poly(vinylpolypyrrolidone) (Sigma-Aldrich, catalog number: P-6755 )
- Ammonium sulfate [(NH4)2SO4] (AppliChem GmbH, Panreac, catalog number: 141140 )
- dH2O (deionized water) (processed by the Milli-Q system from Merck Millipore Corporation)
- Coomassie Blue-stained SDS-PAGE (optional)
- Extraction buffer (EB) (see Recipes)
- Sucrose gradient buffer (SGB) (see Recipes)
- Sucrose density gradient (see Recipes)
- Chromatography buffer A (CBA) (see Recipes)
- Chromatography buffer B (CBB) (see Recipes)
- Activation buffer (AB) (see Recipes)
Equipment
- Ultrasonic processor (Sonics Vibracell, model: VCX 500 ) equipped with a 19 mm high-gain probe
- Optical microscope (Microscope Central, Nikon, model: Alphaphot 2 YS2 )
- Magnetic stirrer (Bibby Scientific, Stuart, model: SB161-3 ) and magnetic bars
- Preparative centrifuge (GMI, Beckman Coulter, model: J2-HS ) equipped with a fixed-angle rotor (Beckman Coulter, model: JA-20 )
- Ultracentrifuge (GMI, Beckman Coulter, model: L-70 ) equipped with a fixed-angle rotor (Beckman Coulter, model: 55.2 Ti )
- Two-chamber (2 x 8 ml) gradient mixer (homemade)
- UV Monitor (GE Healthcare, Amersham-Pharmacia Biotech, model: UV-1 with Rec112 register )
- Fast Performance Liquid Chromatography equipment (FPLC) (GE Healthcare, Amersham-Pharmacia Biotech, model: Äkta Prime )
- Peristaltic pump (GE Healthcare, Pharmacia, model: P-1 )
- UV-V spectrophotometer (Shimadzu Scientific Instruments, model: UV-1603 )
- Water jet vacuum pump (aspirator) (KARTELL SPA VIA DELLE INDUSTRIE, model: 1395 )
- Ultrasonic water bath (Scientific Support, Branson, model: 2200 )
Procedure
- Extraction and ammonium sulfate precipitation of Rubisco
- Rubisco can be purified either from freshly harvested cultures (pelleted by centrifugation at 2,000 x g for 5 min) or from frozen pellets (which may be conserved indefinitely if immediately frozen in liquid nitrogen). In the latter case, the frozen cells should be thawed on ice in the presence of extraction buffer (EB) with protease inhibitors (see next step).
- Resuspend about 8 g (wet weight) of cell pellets in 32 ml of ice-cold EB (containing protease inhibitors).
- Sonicate the cell suspension in an ice bath with 6 pulses (of 30 sec each, separated by 60 sec standby intervals) of a 19 mm-high gain probe delivering 100 W. The extent of cell wall breakage may be checked by optical microscopy.
- Add 0.64 g of insoluble polyvinylpolypyrrolidone to the sonicated suspension and stir with a magnetic bar for 5 min in a cold room (4 °C).
- Centrifuge at 25,000 x g for 15 min at 4 °C, collect the supernatant (avoiding the dark green layer on top of the pellet) and measure its volume in a graduated cylinder.
- Add 0.194 g of solid ammonium sulfate per ml of supernatant (this will bring the solution to 35% salt saturation) while stirring with a magnetic bar. Keep stirring for 30 min in the cold room (4 °C).
- Centrifuge at 25,000 x g for 10 min at 4 °C, collect the clear supernatant and measure its volume in a graduated cylinder again.
- Add 0.151 g of solid ammonium sulfate per ml of supernatant (in order to bring the solution to 60% salt saturation) while stirring again for 30 min in the cold room (4 °C).
- Centrifuge at 25,000 x g for 10 min at 4 °C and discard the supernatant. Resuspend the precipitated protein in less than 3 ml of sucrose gradient buffer (SGB) by mixing carefully with a glass rod (do not vortex because this would develop perdurable foam).
- Bring the volume of solution to 5 ml and load it onto two Sephadex G-25 (PD-10) desalting columns (2.5 ml each) previously equilibrated with SGB at 4 °C. Elute each column with 3.5 ml of the same SGB, finally pooling the eluates.
- Rubisco can be purified either from freshly harvested cultures (pelleted by centrifugation at 2,000 x g for 5 min) or from frozen pellets (which may be conserved indefinitely if immediately frozen in liquid nitrogen). In the latter case, the frozen cells should be thawed on ice in the presence of extraction buffer (EB) with protease inhibitors (see next step).
- Sucrose gradient centrifugation
- By using a two-chamber gradient mixer, prepare four plastic thick-wall centrifuge tubes containing a 16 ml linear gradient of 0.2 to 0.8 M sucrose dissolved in SGB.
- Distribute the 7 ml of the desalted eluate on top of the four 16 ml sucrose gradients and balance the tubes for equal weight by pairs.
- Ultracentrifuge the tubes at 132,000 x g for 4 h in a fixed angle rotor at 4 °C. Select a slow initial acceleration to avoid perturbation of the gradient.
- After centrifugation, fractionate the content of each tube using a glass cannula (e.g., a disposable micro pipet) stuck to the bottom of the tube, connected to an aspiring peristaltic pump in line with a UV-monitor adjusted to 280 nm (Figure 1).
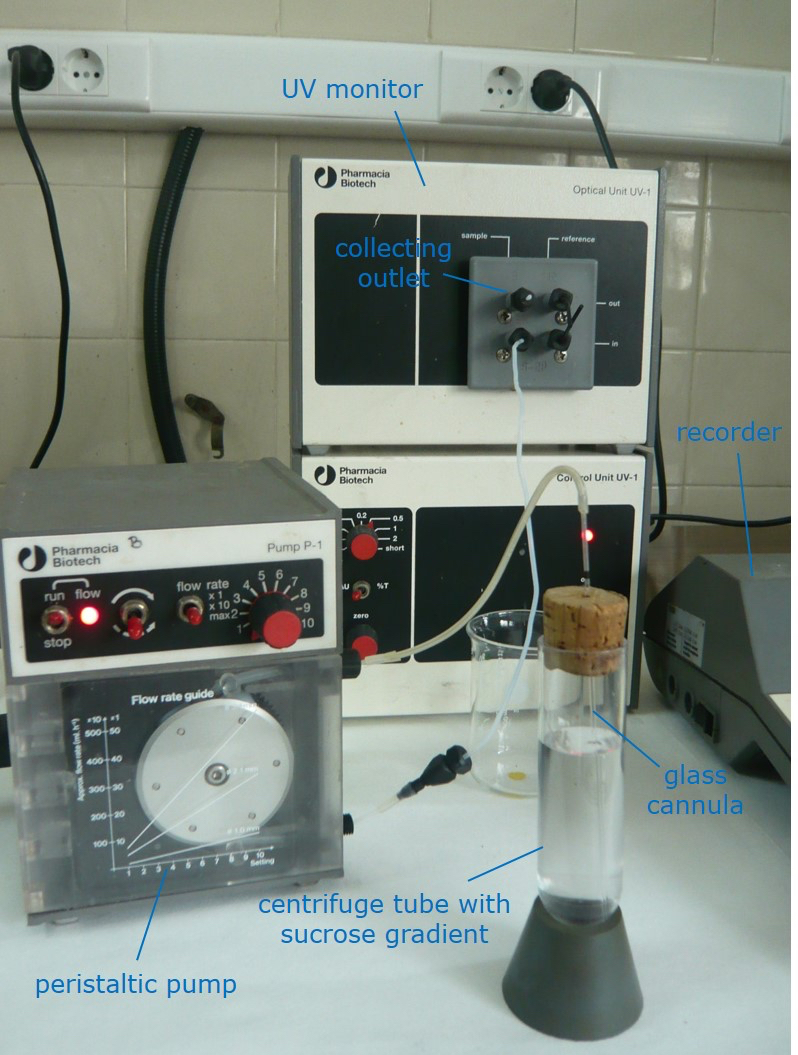
Figure 1. Assembly for collecting fractions from the sucrose gradient after ultracentrifugation - Collect separately the distinct Rubisco peak which appears within 2 and 5 ml from the bottom of the tube (see Figure 2). Combine the collected peaks from the four tubes and store frozen at -20 °C unless the ensuing chromatography is to be performed immediately. Gradient fractions may be stored at -20 °C indefinitely.
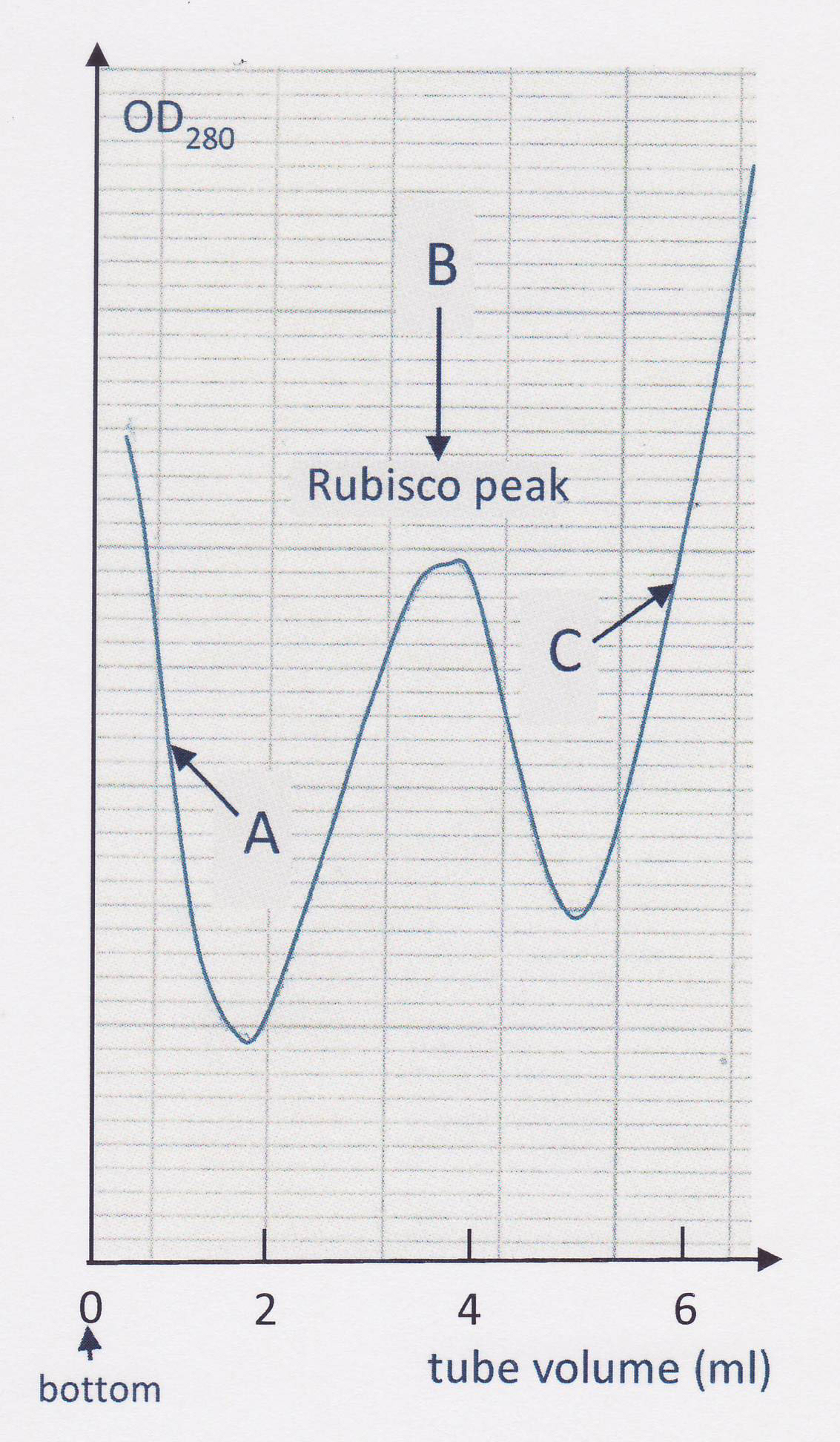
Figure 2. Typical record of the optical density at 280 nm of the sucrose gradient collected from the bottom of the tube after ultracentrifugation. The peak of Rubisco (B) is flanked by a descending absorbance due to nucleic acids (A) and an increasing absorption by the bulk of other (less weighty) proteins (C).
- By using a two-chamber gradient mixer, prepare four plastic thick-wall centrifuge tubes containing a 16 ml linear gradient of 0.2 to 0.8 M sucrose dissolved in SGB.
- Anion-exchange chromatography
- Dilute the pooled Rubisco-containing fractions obtained from the sucrose gradient three-fold with chromatography buffer A (CBA).
- Load the diluted fractions on a 6 ml Resource Q ion-exchange column mounted on a FPLC system and equilibrated with CBA at a 2 ml/min flux rate.
- Elute the column at a 6 ml/min flux with a programmed 120 ml (i.e., 20 column volumes) linear gradient ranging from 0% to 42% of chromatography buffer B (CBB) in CBA while monitoring the absorbance at 280 nm of the eluate. Collect the Rubisco, which will elute as a sharp and very intense peak around a 25% of CBB. Afterwards, the column can be thoroughly washed with 100% CBB, before re-equilibrating it with CBA for processing the next sample. Otherwise, follow the manufacturer's instructions for column storage.
- Dilute the pooled Rubisco-containing fractions obtained from the sucrose gradient three-fold with chromatography buffer A (CBA).
- Rubisco desalting
- Load 2.5 ml of the collected Rubisco solution onto a Sephadex G-25 (PD-10) desalting column equilibrated with activation buffer (AB) and elute the protein with 3.5 ml of the same buffer. This step will eliminate salts [which inhibit Rubisco activity at concentrations higher than 0.2 M (Sudhani et al., 2013)], and provide Mg2+ and CO2, which promote the catalytically competent conformation of Rubisco.
Representative data
The final preparation should contain more than 95% of Rubisco protein being substantially free of nucleic acids (A280/A260 ratio higher than 1.9). Rubisco concentration may be calculated from the UV absorbance assuming an extinction coefficient of ε1% = 15.9 dl·g-1·cm-1. at 280 nm. Typical final concentration of Rubisco is about 1 mg/ml.
Figure 3 illustrates the degree of purity as ascertained by Coomassie Blue-stained SDS-PAGE. The described protocol leads to high purity Rubisco that might be used even for crystallization of the enzyme. In case that such a degree of purity is not needed, the protocol might be shortened omitting the anion exchange chromatography (section C). After the sucrose gradient centrifugation, Rubisco is already more than 80% of the total protein in the extract (see Figure 3B), although there is still a significant contamination of nucleic acids.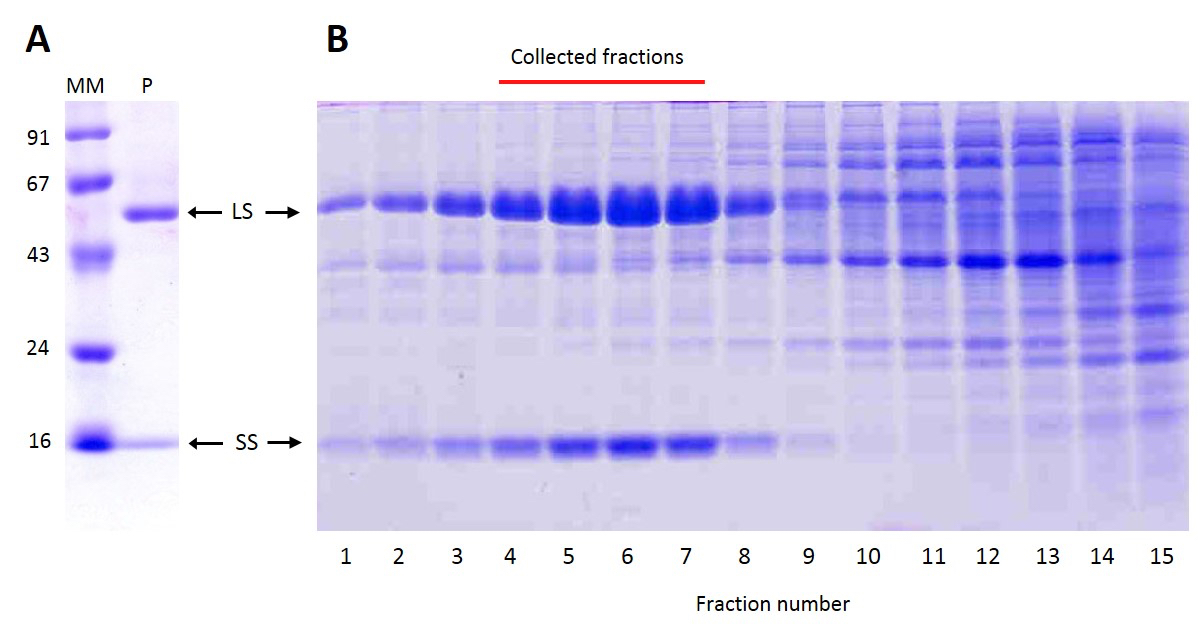
Figure 3. Coomassie-stained SDS-PAGE analysis of Rubisco purification from C. reinhardtii. Arrows indicate the position of the large (LS) and small (SS) subunits of Rubisco on a 14% acrylamide SDS-PAGE gel. A. Final preparation of purified Rubisco (0.25 μg, lane P) run in parallel to markers (MM) of molecular mass (in kDa). B. Fractions (0.7 ml each) collected from the bottom of the sucrose gradient after centrifugation. The red line above indicates the fractions to be pooled for further purification by anion exchange chromatography.
Notes
- We have successfully tested the same purification protocol starting from crude extracts obtained from many other species including leaves of higher plants (spinach, rice, mulberry and orange trees), fronds of Lemna, and cultures of Euglena. Only when purifying Rubisco from diatom species, the ammonium sulfate cuts had to be shifted (to 0-35% saturation) for better yield (Marín-Navarro et al., 2010).
Recipes
- Extraction buffer (EB) (100 mM Tris-sulfate, 10 mM MgSO4, 20 mM 2-mercaptoethanol, pH 8.0)
For 200 ml:
Dissolve 2.42 g of Tris and 0.493 g of MgSO4.7H2O in about 150 ml of dH2O
Adjust the pH to 8.0 with diluted (e.g., 0.5 M) H2SO4
Bring the final volume to 200 ml and store at 4 °C
Dissolve 1 tablet of protease inhibitors per 50 ml, and add 1.4 μl of 2-mercaptoethanol per ml immediately before use. - Sucrose gradient buffer (SGB) (10 mM Tris-H2SO4, 10 mM MgSO4, 10 mM NaHCO3, 1 mM β-mercaptoethanol, pH 8)
For 200 ml:
Dissolve 2.42 g of Tris, 0.493 g of MgSO4.7H2O and 0.168 g of NaHCO3 in about 150 ml of dH2O
Add 140 μl of 2-mercaptoethanol
Adjust the pH to 8.0 with diluted (e.g., 0.5 M) H2SO4
Bring the final volume to 200 ml
This buffer should be prepared fresh for each experiment shortly before dissolving the sucrose and preparing the gradients. - Preparing the sucrose density gradient
For 4 gradients:
0.2 M sucrose solution: Dissolve 2.55 g of sucrose in 35 ml of SGB
0.8 M sucrose solution: Dissolve 9.30 g of sucrose in 28.2 ml of SGB
Fill the mixing (stirred) chamber of a gradient mixer with 8 ml of the 0.8 M sucrose solution and the auxiliary chamber with 8 ml of 0.2 M sucrose solution.
Connect the two chambers and direct the outflow through a peristaltic pump to a thick wall ultracentrifuge tube, letting the mixed solution drop along the wall to the bottom of the tube (see Figure 4).
Repeat this procedure four times to get four gradients in four separate centrifuge tubes.
Cool down the gradients to 4 °C by leaving them undisturbed (i.e., no vibrations) in the cold room for several hours or overnight.
Figure 4. Preparing the sucrose density gradients - Chromatography buffer A (CBA) (20 mM Tris-chloride, pH 7.5)
For 1 L:
Dissolve 2.42 g of Tris in about 0.8 L of dH2O
Carefully adjust the pH to 7.5 using diluted (e.g., 0.2 M) HCl
Bring the volume to 1 L and filter through a sterilizing 0.22 μm disposable filtration system connected to an aspirator vacuum pump
Shortly before using, degas the buffer by placing the bottle (losely open) in an ultrasonic water bath for 10 min - Chromatography buffer B (CBB) (20 mM Tris-chloride, 1 M NaCl, pH 7.5)
For 0.5 L:
Dissolve 1.21 g of Tris and 29.58 g of NaCl in about 0.4 L of dH2O
Carefully adjust the pH to 7.5 using diluted (e.g., 0.2 M) HCl
Bring the volume to 0.5 L, and filter and degas as for CBA - Activation buffer (AB) (100 mM Tris-HCl, 10 mM MgCl2, 10 mM NaHCO3, pH 8.2)
For 100 ml:
Dissolve 1.210 g of Tris, 0.203 g of MgCl2.6H2O and 0.084 g of NaHCO3 in some 80 ml of dH2O
Adjust the pH to 8.2 using diluted (e.g., 1 M) HCl
Bring the final volume to 100 ml and transfer to a 100 ml bottle leaving little headspace. This buffer should be prepared shortly before use to avoid loss of bicarbonate as CO2.
Acknowledgments
This work was supported by a grant (UV-INV-AE14-269247) of the University of Valencia. The protocol is a detailed and expanded version of the one published as supplementary information in Sudhani and Moreno (2015).
References
- Goldwaithe, J. and Bogorad, L. (1975). Ribulose-1,5-diphosphate carboxylase from leaf. Meth Enzymol 42: 481-487.
- Marín-Navarro, J., García-Murria, M. J. and Moreno, J. (2010). Redox properties are conserved in Rubiscos from diatoms and green algae through a different pattern of cysteines. J Phycol 46: 516-524.
- Sudhani, H. P. and Moreno, J. (2015). Control of the ribulose 1,5-bisphosphate carboxylase/oxygenase activity by the chloroplastic glutathione pool. Arch Biochem Biophys 567: 30-34.
- Sudhani, H. P., Garcia-Murria, M. J. and Moreno, J. (2013). Reversible inhibition of CO2 fixation by ribulose 1,5-bisphosphate carboxylase/oxygenase through the synergic effect of arsenite and a monothiol. Plant Cell Environ 36(6): 1160-1170.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sudhani, H. P. K., García-Murria, M. J., Marín-Navarro, J., García-Ferris, C., Peñarrubia, L. and Moreno, J. (2015). Purification of Rubisco from Chlamydomonas reinhardtii. Bio-protocol 5(23): e1673. DOI: 10.21769/BioProtoc.1673.
Category
Plant Science > Phycology > Protein > Isolation and purification
Plant Science > Plant biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link















