- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Single Molecule RNA FISH in the Mammalian Oocyte
Published: Vol 5, Iss 23, Dec 5, 2015 DOI: 10.21769/BioProtoc.1666 Views: 11286
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Visualization of RNA at the Single Cell Level by Fluorescent in situ Hybridization Coupled to Flow Cytometry
Alice Coillard and Elodie Segura
Jun 20, 2018 7265 Views

Evaluation of Genotoxicity by Micronucleus Assay in vitro and by Allium cepa Test in vivo
Christina N. Banti and Sotiris K. Hadjikakou
Jul 20, 2019 9356 Views

SIRF: A Single-cell Assay for in situ Protein Interaction with Nascent DNA Replication Forks
Sunetra Roy and Katharina Schlacher
Sep 20, 2019 8072 Views
Abstract
RNA fluorescence in situ hybridization is a method to localize and measure gene expression in individual cell or tissue. Using multiple specific fluorescently labeled oligonucleotides greatly increases signal-to-noise ratio and thus enables detection of single RNA molecule. Around forty different DNA oligonucleotides designed to common RNA target and labeled with single fluorophore at 3´ terminus hybridizes with target RNA in fixed cells. We adapt this method to visualize target RNA in the mammalian oocyte. The ability to detect single transcript in the mammalian oocyte was challenging due to its large cell size. This method consists of four simple steps: fixation, permeabilization, hybridization and imaging. The protocol is adapted to this large nonattached cell to visualize maternal RNAs.
Combination of various fluorophores allows detection of more RNA targets. This method might be used with organelle markers or expanded with immunofluorescence protocol.
Materials and Reagents
- Cover glasses thickness No.1 22 x 22 mm (Marienfeld-Superior, catalog number: 0101050 )
- Fisherfinest Premium Microscope Slides (Thermo Fisher Scientific, catalog number: 125447 )
- Tissue culture 24-well plates (TPP Techno Plastic Products AG, catalog number: 92024 )
- Tissue culture 96-well plates (TPP Techno Plastic Products AG, catalog number: 92096 )
- Borosilicate glass capillary 3.3. (Hilgenberg GmbH, catalog number: 1409363 ), hand pipette for manipulation) with tip diameter around 100 µm (Hilgenberg GmbH)
- Microtube from Eppendorf 15 ml RNase free (Sigma-Aldrich, catalog number: 0030123.328 )
Note: Pricing & availability is not currently available. - Aluminum foil (29 cm x 150 m) (MAIS.s.r.o.)
- Oocytes from Six-weeks-old female CD1 mice; isolation of mouse oocytes described in Bio-protocol (Tetkova and Hancova, 2015), also (Susor et al., 2015)
- Paraformaldehyde final concentration 4% in PBS (store at -20 °C) (Thermo Fisher Scientific, Afla Aesar, catalog number: 30525894 )
Note: Currently, it is “Sigma-Aldrich, catalog number: 30525894 ”. - Triton X-100 final concentration 2% in 1x PBS (fresh) (Sigma-Aldrich, catalog number: 9002931 )
- 20x saline-sodium citrate (SSC) stock is diluted in RNase free water, in the same day of experiment (Sigma-Aldrich, catalog number: S6639 )
- Ethanol 100% (for final concentration 70% ethanol, freshly dilute 100% ethanol with DEPC water) (Merck Millipore Corporation, catalog number: 1085430250 )
- VECTASHIELD HardSetTM with DAPI (storage 2-8 °C) (Vector Laboratories, catalog number: H1500 )
- ProtectRNA RNase Inhibitor 500x concentrated (final working concentration is 1x; storage at 2-8 °C in dark) (Sigma-Aldrich, catalog number: R7397 )
- Stellaris probes (stock concentration 5 nmol) (Bioresearch Technologies)
- Nuclease-free water (Life Technologies, Ambion®, catalog number: AM9932 )
Note: Currently, it is “Thermo Fisher Scientific, AmbionTM, catalog number: AM9932”. - Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P4417 )
- Poly(vinyl alcohol) (PVA) (Sigma-Aldrich, catalog number: 341584 )
- Formamide (5 ml for 10% final concentration) (Sigma-Aldrich, catalog number: F7503 )
- Dextran sulfate sodium salt from Leuconostoc spp. (Sigma-Aldrich, catalog number: D8906 )
- Wash buffer (see Recipes)
- Hybridization buffer (see Recipes)
- Isolation buffer PVA (see Recipes)
Equipment
- Stereomicroscope Zeiss 2000C (Thermo Fisher Scientific)
- Heating plate (P-lab)
- Multi Bio 3D mini-Shaker (Biosan Laboratories)
- Thermostat incubator mini I5110 (Labnet International)
- Confocal microscope Leica SP5 (Leica Microsystems)
- Filter sets appropriate for fluorophores and excitation lasers
- 40x or 63x or 100x oil immersion objective
Procedure
- Fixation and permeabilization of oocytes
- Wash isolated oocytes in 400 µl isolation buffer PVA and then transfer oocytes into 400 µl of 4% PFA for 25 min room temperature.
- Transfer oocytes into a 24-well dish with 500 µl/well of 2% Triton X-100 with 1x ProtectRNA RNase Inhibitor for 10 min at room temperature.
- Instead of step A2, permeabilize oocytes at room temperature with 70% ethanol for 20 min.
Note: Perform hybridization directly after fixation.
- Wash isolated oocytes in 400 µl isolation buffer PVA and then transfer oocytes into 400 µl of 4% PFA for 25 min room temperature.
- Preparing probe and hybridization
Notes:- For the first use of Stellaris probes, it is recommended to start 4 separate hybridization reactions by adding 1 μl each of 1:50, 1:100, 1:200, 1:400 working dilutions of probes to see which one is optimal. Negative control eGFP, no probe, Malat 1 (Figure 1).
- Warm hybridization buffer to room temperature and mix well. Spin 25 µM working stock probes and dilute to 100 nM 200x times to final concentration into hybridization buffer.
- 500 µl of 37 °C preheated wash buffer with 1x protect RNA RNase inhibitor used for rehydration cells for 15 min.
- Transfer oocytes into 96 well plate with 100 µl/per well hybridization buffer and add probe.
- Protect 96 well plate from light by aluminum foil, incubate in thermostat incubator overnight at 37 °C.
Note: Shaking is not an essential step, it might be omitted.
- For the first use of Stellaris probes, it is recommended to start 4 separate hybridization reactions by adding 1 μl each of 1:50, 1:100, 1:200, 1:400 working dilutions of probes to see which one is optimal. Negative control eGFP, no probe, Malat 1 (Figure 1).
- Post hybridization washes and mounting
- Wash 1 time in 150 µl 2x SSC for 5 min, shake 30 RPM and turning angle 360° at 30 sec of 96 well plate.
Transfer oocytes into 96 well plate with 125 µl/per well of warm wash buffer. Incubate at least 50 min at 37 °C.
Note: Shaking is not an essential step, it might be omitted. - Rinse 1 time in 150 µl of 2x SSC for 2 min at room temperature.
- Put oocytes with 0.5 µl of SSC liquid on glass slide with 20 µl of Vectashiled-hardening mounting medium with DAPI, wait 1 min then cover with cover slip.
- Wash 1 time in 150 µl 2x SSC for 5 min, shake 30 RPM and turning angle 360° at 30 sec of 96 well plate.
- Image sample
- Leave the slide in the dark chamber for 2 h at 4 °C.
- Next, scan with confocal laser scanning microscope.
- Take images in the same day to avoid sample drying and fluorophore fading.
- Leave the slide in the dark chamber for 2 h at 4 °C.
- Imaging setup
Typical settings for oocytes have two channels (red, blue) and objective HCX PL APO lambda 63.0 x 1.25 OIL UV. Another setup: For visualization of Cal flour 635 labeled probes used 633 laser the 618 nm excitation and emission 635 nm wavelength, laser power set at 10%, scan speed 100 Hz, format of image 512 x 512, pine hole 110 μm, Z-stack thickness: 10 μm and image analysis and merging by LAS AF.
Note: To be able to compare images, make sure settings are identical between different experiments (e.g., laser intensity, gain, offset, scan speed, etc.).
Negative control for hybridization without probe.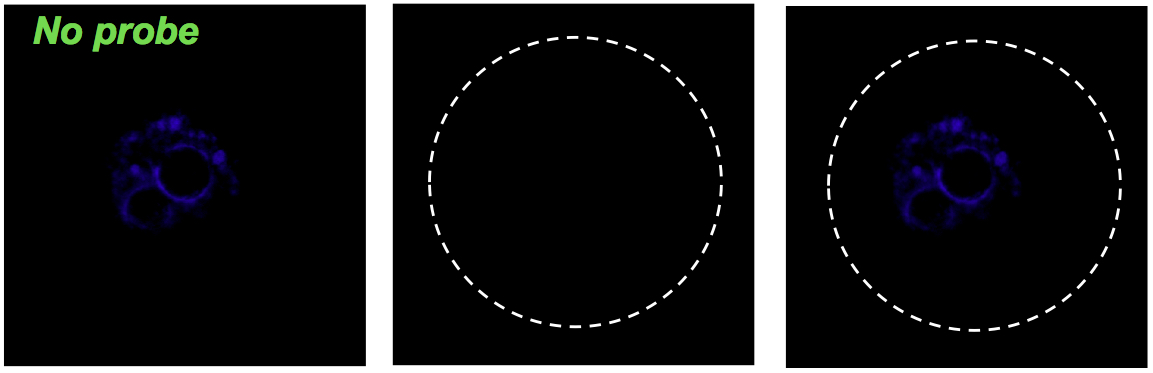
Negative control of experiment. Oocyte without eGFP mRNA (green).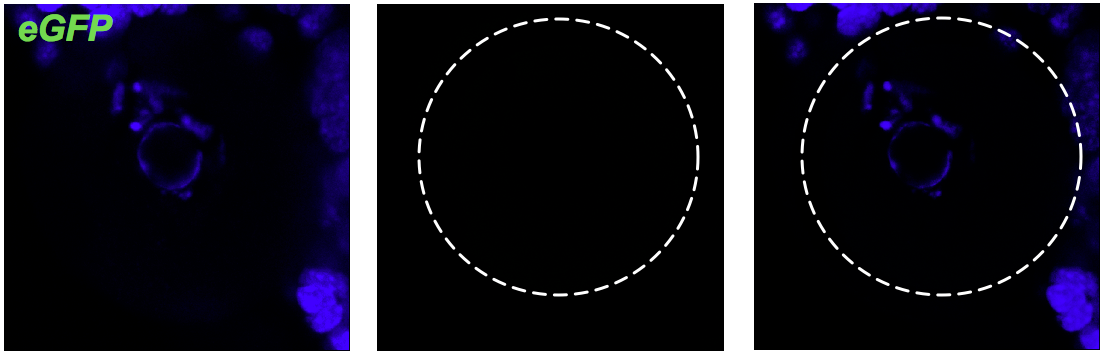
Specific signal of Malat1 (green) in cumulus cells in high probe concentration (1: 50). Oocyte shows high background.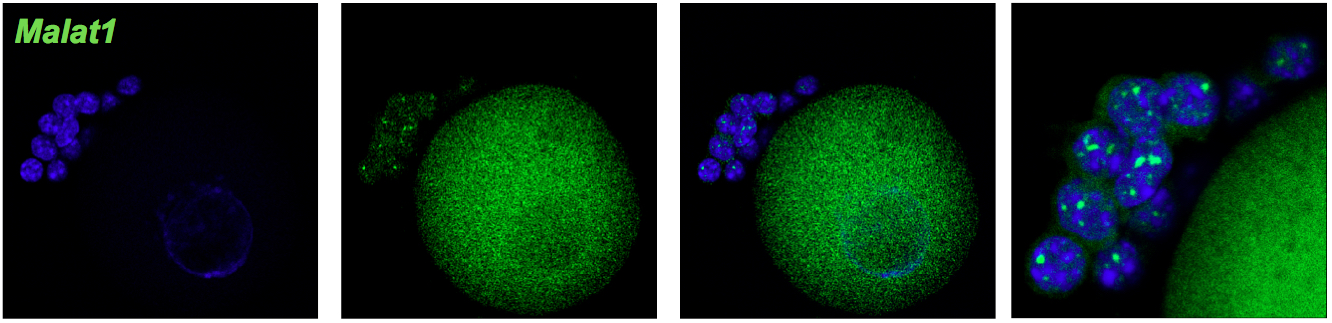
Figure 1. Optimization of FISH protocol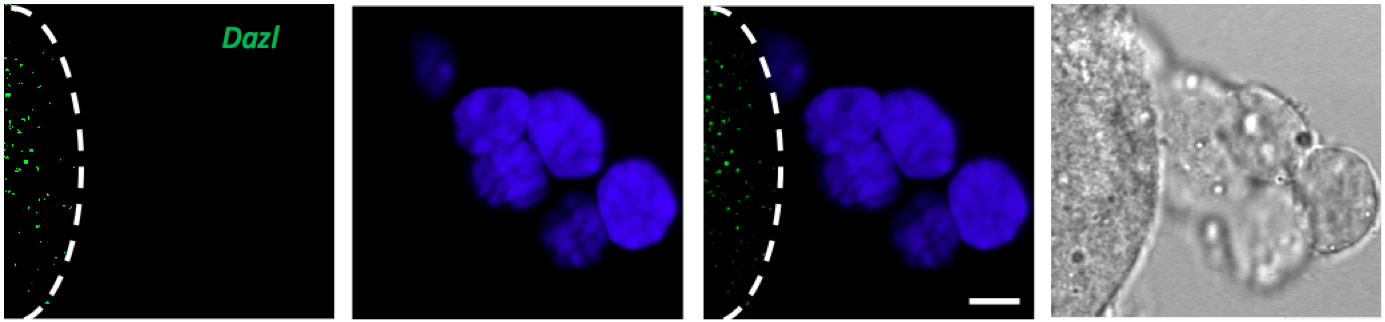
Figure 2. Only oocyte express Dazl mRNA however cumulus cells shows no green signal after FISH against Dazl mRNA. Line marks the edge of the oocyte, DAPI (blue), scale bar 10 µm.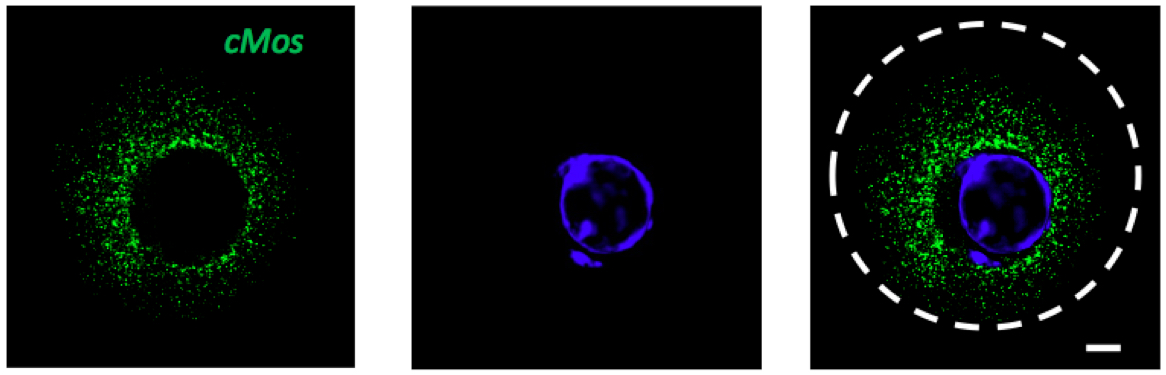
Figure 3. GV stage of mouse oocyte with green signal for cMos mRNA. DAPI (blue), scale bar 10 µm.
Recipes
- Wash buffer (50 ml)
20x SSC (5 ml)
Formamide (5 ml for 10% final concentration)
Nuclease-free water (to 50 ml final volume)
Note: Divide volume into 5 ml aliquots, freeze and stored at -20 °C. - Hybridization buffer (10 ml)
Dextran sulfate (1 g)
20x SSC (1 ml)
Formamide (1 ml for 10% final concentration) RNAse free, stored at 2-8 °C
Nuclease-free water (to 10 ml final volume)
Note: Divide volume into 0.5 ml aliquots, freeze and stored at -20 °C. - Isolation buffer PVA (50 ml)
Phosphate buffer solution (50 ml final volume)
PVA 2 mg
Protect RNA RNase inhibitor 1x concentrate
Note: Wash buffer with the same percentage of formamide as hybridization buffer.
Acknowledgments
This work was supported by research grants 1) Charles University in Prague, Faculty of Cell Biology-GAUK 243-227026 and 2) GACR 13-12291S. Some principles of the described protocol have been adapted from Singer Lab Protocol: Published 1998 and from Christian Lanctot Protocol. The original work was published in Susor et al. (2015).
References
- Bioresearch Technologies: Design and ordering single molecule FISH probe.
- Femino, A. M., Fay, F. S., Fogarty, K. and Singer, R. H. (1998). Visualization of single RNA transcripts in situ. Science 280(5363): 585-590.
- Flemr, M. and Svoboda, P. (2011). Ribonucleoprotein localization in mouse oocytes. Methods 53(2): 136-141.
- Susor, A., Jansova, D., Cerna, R., Danylevska, A., Anger, M., Toralova, T., Malik, R., Supolikova, J., Cook, M. S., Oh, J. S. and Kubelka, M. (2015). Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway. Nat Commun 6: 6078.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jansova, D. (2015). Single Molecule RNA FISH in the Mammalian Oocyte. Bio-protocol 5(23): e1666. DOI: 10.21769/BioProtoc.1666.
Category
Cell Biology > Cell staining > Nucleic acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










