- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Sample Preparation and Fractionation of Arabidopsis thaliana Sperm and Vegetative Cell Nuclei by FACS
Published: Vol 5, Iss 22, Nov 20, 2015 DOI: 10.21769/BioProtoc.1664 Views: 13679
Reviewed by: Samik BhattacharyaYurong XiePengpeng Li

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Isolation of Lysosome-Related Organelles from Stationary Phase and Iron-Overloaded Chlamydomonas reinhardtii Cells
Jiling Li and Huan Long
Nov 20, 2024 1767 Views

Isolation and Biophysical Characterization of Extracellular Vesicles From Hairy Root Cultures
Marisa Conte [...] Alfredo Ambrosone
Mar 5, 2025 2218 Views

Rapid Miniprep of Intact Chloroplasts from Arabidopsis thaliana Leaves
Brenda A. Carranza-Correa [...] Manuel Gutiérrez-Aguilar
May 20, 2025 2645 Views
Abstract
One of the major topics in plant and animal biology is sexual reproduction. It is, therefore, of great interest to isolate and study germ cells and accessory cells. The male gametophyte of the flowering plant Arabidopsis thaliana (A. thaliana), pollen, is the product of two post-meiotic mitotic divisions. Each mature pollen grain consists of two sperm cells contained within the vegetative cell, the non-reproductive companion cell. The tough pollen wall and its special nested structure make it difficult to study pollen cells separately. Here, we describe a simple and efficient method to fractionate A. thaliana sperm and vegetative cell nuclei by fluorescence activated cell sorting (FACS). Our protocol is based on differences in fluorescence intensity of sperm and vegetative cell nuclei stained with SYBR Green I. 100 plants yield about 1 x 106 sperm and 350,000 vegetative cell nuclei. This method can be used for purifying pollen nuclei of various A. thaliana wild-type accessions and mutant lines, and can, in principle, be adapted for pollen of other plant species.
Keywords: SYBR GreenMaterials and Reagents
- Plastic ware / consumables
- 15 ml polypropylene tubes (SARSTEDT AG & Co, catalog number: 62554502 )
- 50 ml polypropylene tubes (SARSTEDT AG & Co, catalog number: 62548004 )
- 100 µm nylon mesh (Biologix Group Limited, catalog number: 151100 )
- 40 µm nylon mesh (Biologix Group Limited, catalog number: 151040 )
- 250 ml centrifuge tubes (Bartelt, catalog number: 9.315 721 )
- 1.5 ml and 2 ml microliter reaction tubes
- Syringe filters 0.2 µm (Bartelt, catalog number: 9055511 )
- BD Falcon® 12 x 75 mm tube with cell strainer cap (BD Biosciences, catalog number: 352235 )
Note: Currently, it is “Corning Inc., catalog number: 352235”. - Sterican disposable hypodermic needles with a diameter of 0.45 x 25 mm (B. Braun Melsungen AG, Braun Petzold, catalog number: 4657683 )
- Pipettes tips (1,000 µl, 200 µl, 20 µl, 2 µl)
- Acid-washed glass beads (see Recipes)
- 15 ml polypropylene tubes (SARSTEDT AG & Co, catalog number: 62554502 )
- Glassware
- Glass beads 0.4-0.6 mm (Sartorius AG, catalog number: BBI8541701 )
- 250 ml Erlenmeyer flask
- Glass tubes
- Glass slides
- Cover slips
- Glass beads 0.4-0.6 mm (Sartorius AG, catalog number: BBI8541701 )
- Plant material
- 100 Flowering wild-type Arabidopsis thaliana plants (Col-0 accession and dml-3 mutant, are used as examples) (Arabidopsis, catalog number: SALK_056440 )
- Chemicals
- D(+)-Sucrose for analysis (AppliChem GmbH, catalog number: 131621.0914 )
- DAPI (4', 6-Diamidino-2-Phenylindole, dihydrochloride) (Life Technologies, InvitrogenTM, catalog number: D1306 )
Note: Currently, it is “Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D1306”. - SYBR Green I nucleic acid stain stock solution (Life Technologies, catalog number: S7567 )
Note: Currently, it is “Thermo Fisher Scientific, InvitrogenTM, catalog number: S7567”. - Phenylmethanesulfonyl fluoride (Sigma-Aldrich, catalog number: 93482 )
- Complete, EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics, catalog number: 11873580001 )
- Calcium chloride dihydrate (AppliChem GmbH, catalog number: A3587 )
- Ethylenediamine tetraacetic acid disodium salt dihydrate (Carl Roth GmbH + Co., catalog number: 8043 )
- Ethylene glycol-bis (2-aminoethylether)-N, N, N, N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- Ficoll® PM 400 (Sigma-Aldrich, catalog number: F4375 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Magnesium acetate tetrahydrate (Sigma-Aldrich, catalog number: M5661 )
- Sodium chloride (EMD Millipore Corporation, catalog number: 106404 )
- Potassium chloride (Sigma-Aldrich, catalog number: P3911 )
- D-Sorbitol (Sigma-Aldrich, catalog number: S6021 )
- Spermine [N, N'-Bis-(3-aminopropyl)-1, 4-diaminobutan] (AppliChem GmbH, catalog number: A0672.0005 )
- TritonTM X-100 (Sigma-Aldrich, catalog number: 234729 )
- Tris-base (AppliChem GmbH, catalog number: A1086 )
- Nitric acid 65% (Carl Roth GmbH + Co., catalog number: X898.1 )
- Hydrochloric acid fuming 37% (Carl Roth GmbH + Co., catalog number: 4625.1 )
- Sodium Hydroxide pellets (AppliChem GmbH, catalog number: A3910 )
- Ultrapure water (MilliQ) (conductivity > 18 MΩ-1 cm-1)
- D(+)-Sucrose for analysis (AppliChem GmbH, catalog number: 131621.0914 )
- Buffers
- BD FACSFlow Sheath Fluid (BD Bioscience, catalog number: 342003 )
- Sucrose solution (see Recipes)
- Buffer A (see Recipes)
- Buffer B (see Recipes)
- Protease inhibitor stock solution (see Recipes)
- DAPI stock solution (see Recipes)
- BD FACSFlow Sheath Fluid (BD Bioscience, catalog number: 342003 )
Equipment
- Instruments
- Beckman Coulter Avanti J-26XP centrifuge with the JA-10 rotor
- Table-top centrifuge
- Orbital shaker (VWR International, catalog number: 89032102 )
- Drying oven (EHRET GmbH, catalog number: 3108 )
- Bead-beater (Retsch GmbH, model: MM301 )
- Fluorescence microscope with 20x, 40x, and 100x objectives and a DAPI filter
- Flow cytometer: FACS Aria I (BD Biosciences) with a 70 µm nozzle and 70 psi sheath pressure, and a 488 nm coherent sapphire solid state 13 mW laser for excitation and a 530/30 nm band-pass filter for detection of SYBR green I
- Beckman Coulter Avanti J-26XP centrifuge with the JA-10 rotor
- General lab equipment
- Beaker
- 250 ml Erlenmeyer flask
- Glass tubes
- Sharp-edged scissors
- Forceps
- Air displacement pipettes
- Ice container
Software
- Flow cytometric software (BD Biosciences, model: FACS Diva 6.1.2)
Procedure
- Release of A. thaliana pollen from inflorescences [as previously described (Honys and Twell, 2003; Johnson-Brousseau and McCormick, 2004; Schoft et al., 2009) with modifications]
- ~100 plants (A. thaliana) were grown for 5 weeks at 16 h light/8 h dark cycles at 22 °C. Developmental stage of the plants corresponds to 6.10 to 6.50 according to Table 2 in Boyers et al. (2001).
- All inflorescences were collected in a beaker (placed on ice) by cutting them off with sharp-edged scissors.
- 9% ice-cold sucrose solution was poured onto the collected inflorescences until they were covered. The inflorescences were vigorously swirled for one minute to release mature pollen grains.
- The suspension was filtered through a 100 µm nylon mesh into 250 ml centrifugation tubes placed on ice.
- Pollen grains were concentrated by centrifugation at 1,350 x g for 10 min at 4 °C.
- The supernatant was poured back onto the inflorescences, and steps 3-5 were repeated once more.
- The pollen pellets were resuspended in ice-cold buffer A to a final volume of 10 ml and filtered through a 40 µm nylon mesh into a 15 ml Falcon tube.
- Pollen grains were precipitated by centrifugation at 800 x g for 10 min at 4 °C.
- Pollen grains were washed once again with 1 ml of buffer A and collected in a sterile 1.5 ml tube by centrifugation at 5,000 x g for 5 min at 4 °C in a table-top centrifuge. The supernatant was discarded.
Notes:- Collecting inflorescences and releasing pollen takes about 2-3 h.
- The precipitated pollen can be used immediately or can be rapidly frozen in liquid nitrogen and kept at -80 ° C for several years.
- 100 A. thaliana plants (Col-0 ecotype) yield about 100 µl of pollen. Pollen pellet volume is estimated by filling an identical, empty tube with water up to the same level as the pollen and measuring the water volume with an air-displacement pipet. This amount can vary depending on accession, growth conditions, and age of the plant.
- Collecting inflorescences and releasing pollen takes about 2-3 h.
- ~100 plants (A. thaliana) were grown for 5 weeks at 16 h light/8 h dark cycles at 22 °C. Developmental stage of the plants corresponds to 6.10 to 6.50 according to Table 2 in Boyers et al. (2001).
- Isolation of A. thaliana pollen nuclei from mature pollen
- 10-20 sterile 2 ml centrifuge tubes were each filled with 1.8 g of acid-washed glass beads. Unused tubes can be stored at room temperature and used later.
- Harvested pollen were mixed with buffer A supplemented with PMSF and protease inhibitors using the following pollen to buffer A ratios:
Below a pollen pellet volume of 80 µl a ratio of 1:3 (pollen:buffer A) was used between 80 µl and 150 µl a ratio of 1 : 3.5 was used above 150 µl, a ratio of 1 : 4 was used for resuspension of pollen grains in buffer A. - The pollen grain suspension was carefully pipetted into 2 ml centrifuge tubes filled with glass beads (prepared in step B1) in 50 µl aliquots.
- Pollen grains were subjected to bead beating by placing the 2 ml tubes filled with pollen suspension and glass beads in a Retsch ball mill and, subsequently, running a beating protocol for 1.5 min at 15 Hz.
- Three holes were punched into the bottom of the tubes with a fresh needle. Lids were carefully cut off, and the 2 ml tubes were placed on top of sterile 1.5 ml tubes that have also had their lids removed. The two tubes were then placed into glass tubes for centrifugation, so that the nuclei suspension will be recovered in the 1.5 ml tube (alternatively, 15 ml Falcon tubes can be used) (Figure 1A).
- Glass tubes were centrifuged at 800 x g for 10 min at 4 °C. The 2 ml tubes containing the glass beads were discarded and the nuclei suspensions were pooled into a single sterile 1.5 ml centrifuge tube.
- 10 µl of pollen nuclei suspension was mixed with 1 µl of DAPI in water and pipetted onto a glass slide. A cover slip was carefully placed on top of the sample. Efficiency of pollen nuclei release and quality of nuclei were assessed with a fluorescence microscope using 20x and 40x objectives and the DAPI channel [excitation/emission of DAPI (nm): 358/461] (Figure 1B and C).

Figure 1. Assessment of A. thaliana pollen nuclei release efficiency. A. Three-tube setup before and after centrifugation. B. Microscopic image of A. thaliana wild-type (Col-0) pollen before bead beating, DAPI: DAPI-stained pollen examined with the DAPI filter. 40x magnification. Size bar: 25 µm. C. Microscopic image of A. thaliana wild-type pollen (Col-0) after bead beating. Arrows: (1) intact pollen, (2) nuclei still attached to cytoplasm, (3) empty pollen shell, (4) released vegetative nucleus. DAPI: DAPI-stained pollen examined with the DAPI filter. 40x magnification. Size bar: 25 µm
Notes:- Centrifuge tubes should be kept at 4 °C whenever possible.
- The ratio between the volume of pollen and the volume of buffer A is critical for optimal disruption of pollen walls. The ratios described here lead to ~80-90% disruption efficiency. Adjust the ratio if pollen remains intact, or if the sample is heavily damaged as this will result in low yield of nuclei. More buffer increases breaking efficiency (and the risk of damage).
- To optimize the beating protocol we suggest to use two different frequencies and to run the beating protocol for 1, 1.5 and 2 min. 10 µl aliquots of the six samples should be mixed with 1 µl DAPI working solution and inspected at the microscope. 70-90% of all pollen grains should be broken and most of the released nuclei should float in the solution.
- Check the pH of the buffers if released nuclei are swollen and yellowish.
- Isolation of pollen nuclei takes about one hour.
- Centrifuge tubes should be kept at 4 °C whenever possible.
- 10-20 sterile 2 ml centrifuge tubes were each filled with 1.8 g of acid-washed glass beads. Unused tubes can be stored at room temperature and used later.
- Fractionation of A. thaliana sperm and vegetative cell nuclei by fluorescence-activated cell sorting
- The cell sorter was equipped with a 70 µm nozzle and the sheath pressure was adjusted to 70 psi. For excitation, a 488 nm Coherent Sapphire Solid State 13 mW Laser was used. SYBR Green fluorescence was detected by a 530/30 nm band-pass filter. Cooling units were used to keep sample and fractionated nuclei at 4 °C during the whole procedure. Two labelled 15 ml Falcon tubes were placed into the device as collection tubes.
- The nuclei suspension was diluted with 0.5 volumes of buffer B and 250 µl were transferred to FACS tubes by filtering through 35 µm cell strainer caps.
- 5 µl of SYBR Green I was pipetted to one of the samples, mixed, and incubated for 5 min. Additional FACS tubes were kept on ice and stained with SYBR Green I just before sorting.
- Using the FACS Diva 6.1.2 software, a new FACS graph was prepared showing SSC-A (side-scatter area) on the y axis and FITC-A (area) on the x axis, both on a linear scale.
- To begin sorting, an unstained sample was analysed as negative control.
- Next, the SYBR Green I stained nuclei were loaded into the FACS device. Two fluorescent populations appear on the FACS plot, which were separately gated. For gating, a square was drawn around the densest part of the population. Outliers were ignored. Gating along the y-axis is not so critical whereas care has to be taken to avoid the area along the x-axis where both populations overlap to ensure purity of the sorted populations. To avoid sorting aggregates, like two adherent sperm nuclei, an auxiliary graph was set up that only showed the events gated in the first graph. This auxiliary plot with FITC-W (FITC width) on the y axis and FITC-A (FITC area) on the x axis reveals differences in the FITC-W value, which is higher in aggregates than in single nuclei. The two nuclear populations marked in Figure 2 B and D represent the desired nuclei and were gated for sorting. All events in this plot are gated. Once a sample tube was empty, a freshly stained sample tube was loaded into the FACS device.
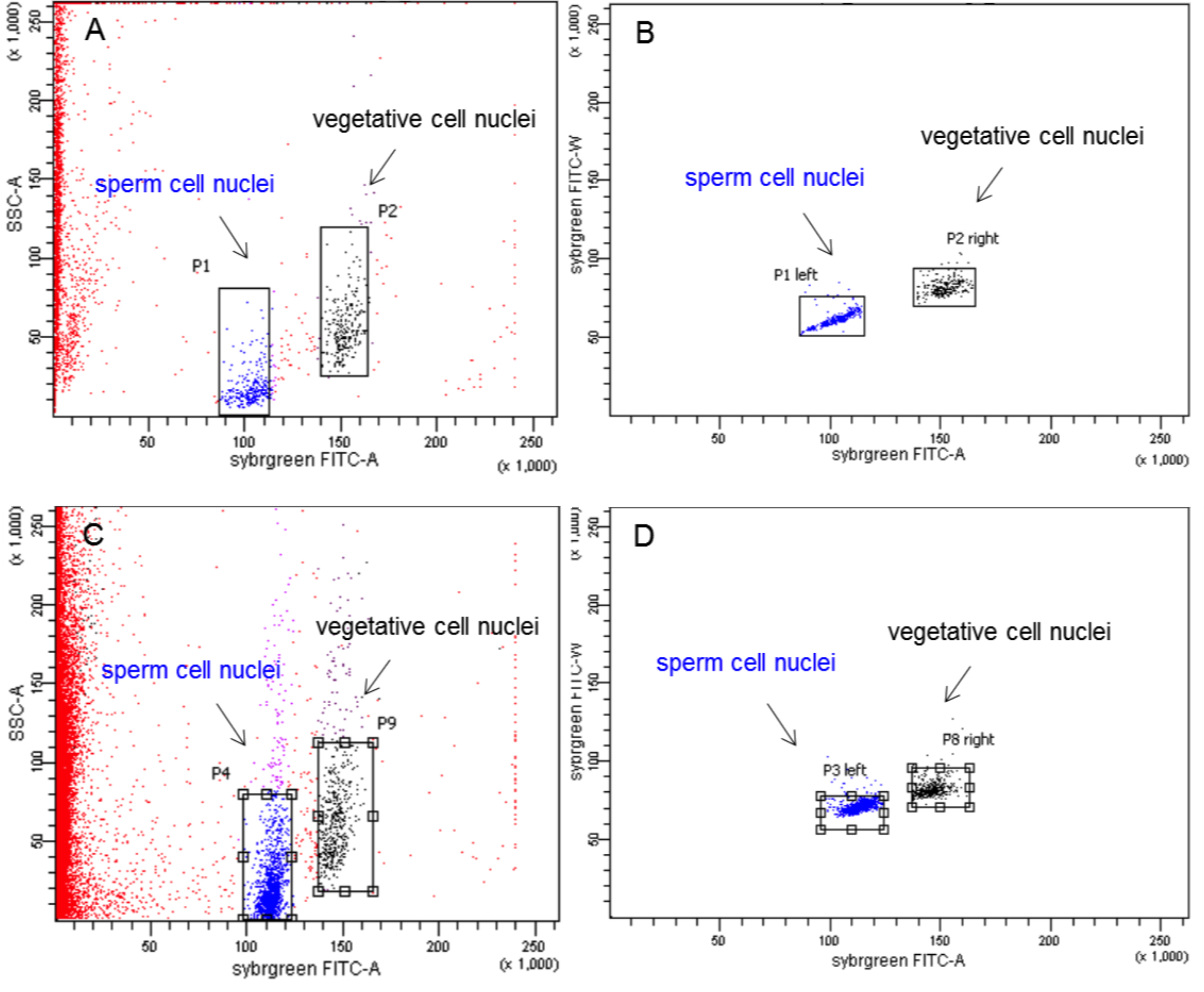
Figure 2. FACS plots of pollen nuclei during sorting. A. Wild-type A. thaliana (Col-0) pollen nuclei displayed as side scatter versus FITC-A (FITC area) of all events on linear scales. B. Auxiliary graph with FITC-W (FITC width) on the y axis and FITC-A (FITC area) on the x axis in linear scales that shows events gated in graph (A). The left population consists of sperm nuclei, the right population represents the vegetative nuclei. Both populations were gated for sorting. C. Dml-3 mutant (SALK_056440) A. thaliana pollen nuclei displayed as side scatter versus FITC-A (FITC area) of all events on linear scales. D. Auxiliary graph for graph (C). - After sorting, a 10 µl aliquot of each population was stained with DAPI and inspected under a fluorescence microscope (as in step B7). Using 100x magnification, sperm and vegetative cell nuclei could be identified by differences in nucleus size, shape, and intensity of DAPI fluorescence (Figure 3). The average diameter (measured through the longest part) of sperm cell nuclei is 2.42 μm (which corresponds to Borges et al., 2012), the average diameter (measured through the longest part) of vegetative cell nuclei is 4.64 μm (Figure 3E). Sperm cell nuclei are regularly shaped (round or ellipsoid) whereas vegetative cell nuclei are highly lobed. DAPI stained sperm nuclei appear brighter than vegetative cell nuclei. The purity of each population was about 99%.
Notes:- The wrong FACS setting will result in no or only one population of nuclei in the FACS plot. Use the right nozzle and high pressure setting.
- Make sure to use a linear scale on both axes of the FACS plot.
- Use more SYBR green I if the nuclei populations in the FACS plot are very close to each other.
- If the population of vegetative cell nuclei is disappearing during the FACS run or if the yield is too low, the buffers and/or plants were too old.
- As the samples are not fixed, populations might move slightly over time and gating has to be monitored and adjusted if necessary.
- Fixing the sample before FACS (e.g., with formaldehyde) will result in large nuclear aggregates and is, therefore, not recommended.
- Depending on the amount of pollen used, FACS will take at least two hours.
- After the sort, rinse the system with diluted bleach to remove all SYBR Green I traces.
- 200 mg (~100 µl) wet mature pollen will result in ~1 x 106 sperm and 350,000 vegetative cell nuclei.
- The expected 2:1 ratio of sperm to vegetative cell nuclei cannot be obtained, as vegetative cell nuclei have a more fragile morphology compared to sperm cell nuclei and can be lost. The average sperm versus vegetative cell nuclei ratio is 3:1.
- After the sort finished, we usually continued with purification. DNA extraction was performed as described in Schoft et al. (2011). RNA extraction was done according to Schoft et al. (2015). The samples can also be fixed (Schoft et al., 2015). Although we never tried, the samples should, in principle, survive flash freezing and storage at -80 °C.
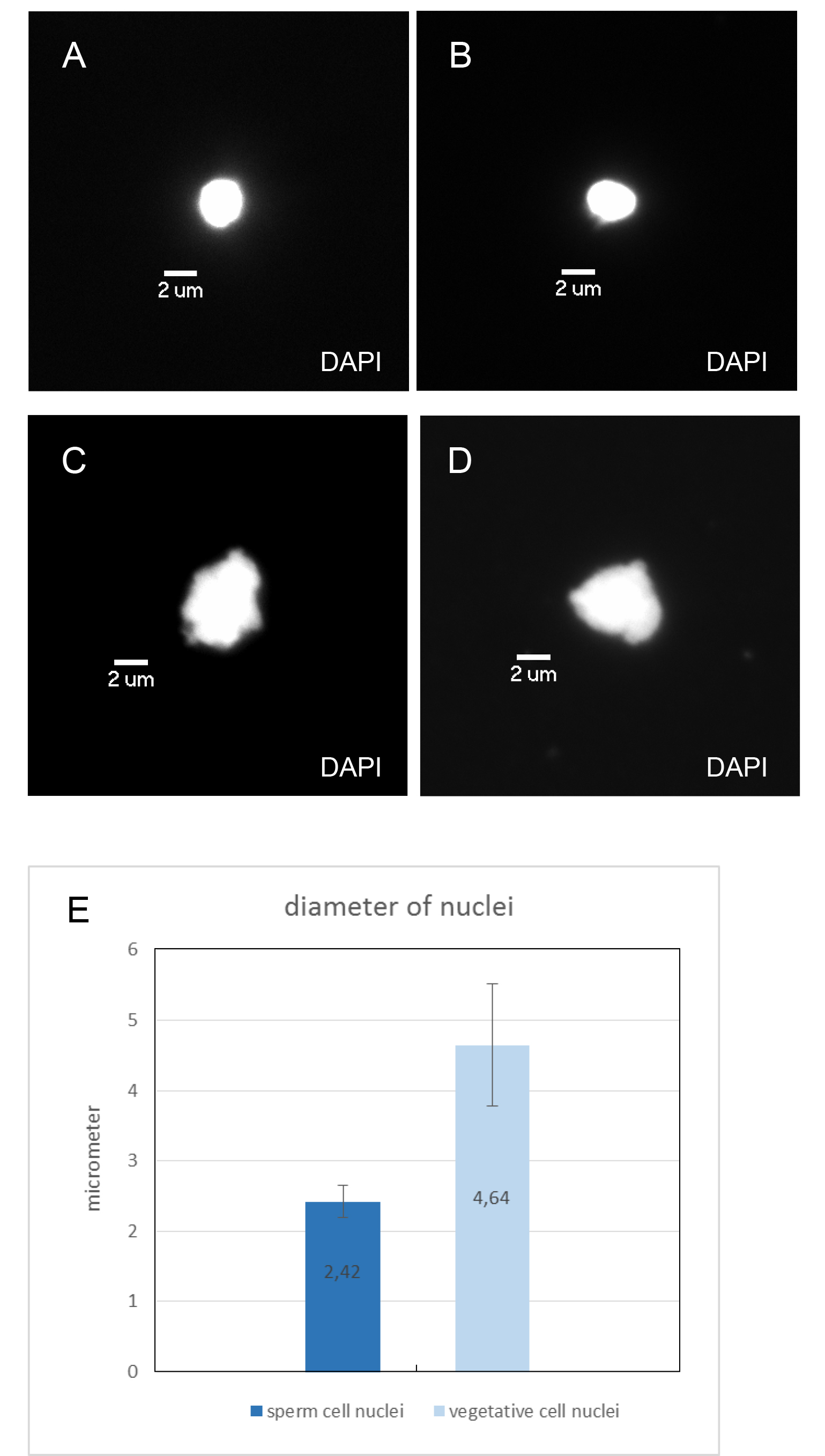
Figure 3. Microscopic images of sorted A. thaliana pollen nuclei. A-D. Nuclei were counterstained with DAPI and examined with the DAPI filter. Magnification 100x, size bar 2 μm. A and B Sperm cell nuclei. C and D Vegetative cell nuclei. E. Average diameter of sorted sperm and vegetative cell nuclei (measured through the longest part of the nuclei; length in μm; error bar: standard deviation).
- The wrong FACS setting will result in no or only one population of nuclei in the FACS plot. Use the right nozzle and high pressure setting.
- The cell sorter was equipped with a 70 µm nozzle and the sheath pressure was adjusted to 70 psi. For excitation, a 488 nm Coherent Sapphire Solid State 13 mW Laser was used. SYBR Green fluorescence was detected by a 530/30 nm band-pass filter. Cooling units were used to keep sample and fractionated nuclei at 4 °C during the whole procedure. Two labelled 15 ml Falcon tubes were placed into the device as collection tubes.
Recipes
- Acid-washed glass beads
- Carefully pour glass beads into a 250 ml Erlenmeyer flask. Don't add more than 100 ml of glass beads.
- Cover the glass beads with nitric acid and shake the flask in an orbital shaker overnight. Use speed settings that allow the glass beads to move a bit.
- Discard the nitric acid and wash the glass beads with water until the pH is neutral.
- Remove the water and sterilize the glass beads in a drying oven at 180 °C for 4 h.
- Carefully pour glass beads into a 250 ml Erlenmeyer flask. Don't add more than 100 ml of glass beads.
- Sucrose solution (working solution 9%, stock 45%)
- Dissolve 450 g sucrose in 1 L of water, autoclave and keep at room temperature.
- Dilute 1:5 with sterile water for working solution.
- Dissolve 450 g sucrose in 1 L of water, autoclave and keep at room temperature.
- Buffer A
- 1 M sorbitol, 7% ficol PM 400, 5 mM MgAc, 3 mM CaCl2, 5 mM EGTA, 50 mM Tris-HCl (pH 7.5), 100% Triton X100, glycerol.
- Mix 9.1 g sorbitol, 3.5 g Ficoll PM 400, 0.05 g Mg acetate, 0.022 g CaCl2, 0.095 g EGTA and 0.3 g Tris in 35 ml of sterile water.
- Adjust the pH to 7.5 with 1 M HCl.
- Add 1 ml Triton x100 (final concentration 2%) and 10 ml glycerol (final concentration 20%). Stored at 4 °C. The solution can be used for 3-4 weeks.
- Protease inhibitor cocktail (1:50 dilution) and PMSF (1:100 dilution) should be added just before use.
- 1 M sorbitol, 7% ficol PM 400, 5 mM MgAc, 3 mM CaCl2, 5 mM EGTA, 50 mM Tris-HCl (pH 7.5), 100% Triton X100, glycerol.
- Buffer B
- 15 mM Tris-HCl (pH 7.5), 2 mM Na2-EDTA, 0.5 mM spermine·4 HCl, 80 mM KCl, 20 mM NaCl, 2% Triton x100.
- Stock solutions:
- For 1 M Tris-HCl, dissolve 6.05 g in 30 ml water, adjust the pH to 7.5 with 5 M HCl and bring up to 50 ml with sterile water, autoclave;
- For 0.5 M EDTA, adjust the pH of 350 ml water to 8 with NaOH pellets and slowly add the salt while stirring, bring water up to 500 ml, autoclave;
- For 1 M spermine, dissolve 0.2 g in 10 ml water (final volume), sterile-filter;
- For 3 M KCl dissolve 22.365 g in 100 ml water (final volume), autoclave;
- For 5 M NaCl dissolve 29.22 g in 100 ml water (final volume), autoclave.
- For 5 ml working solution mix 75 µl Tris-HCl, 20 µl EDTA, 25 µl spermine, 133 µl KCl, 20 µl NaCl and 100 µl Triton X-100, bring up to 5 ml with sterile water.
- Stored at 4 °C.
- Protease inhibitor cocktail (1:50 dilution) and PMSF (1:100 dilution) should be added just before use.
- For 1 M Tris-HCl, dissolve 6.05 g in 30 ml water, adjust the pH to 7.5 with 5 M HCl and bring up to 50 ml with sterile water, autoclave;
- 15 mM Tris-HCl (pH 7.5), 2 mM Na2-EDTA, 0.5 mM spermine·4 HCl, 80 mM KCl, 20 mM NaCl, 2% Triton x100.
- Protease inhibitor stock solution
For 50x concentration, one tablet is dissolved in 1 ml water - DAPI (working solution 10 µg/ml; stock 2 mg/ml)
- Add 5 ml sterile water to 10 mg of DAPI powder for the stock solution
- Dilute stock solution 1:200 with sterile water for the working solution (“DAPI in water”)
- Protect from light
- Stored at -20 °C
- Add 5 ml sterile water to 10 mg of DAPI powder for the stock solution
Acknowledgments
This protocol is based on previously published work (Schoft et al., 2009; Schoft et al., 2011; Ibarra et al., 2012; Schoft et al., 2015). We thank the BioOptics Facility of the Research Institute of Molecular Pathology, Vienna, for setting up and optimizing FACS. We thank Gerald Schmauss for measuring sizes of sorted A. thaliana pollen nuclei. We thank Hisashi Tamaru for his support. This work was supported by Austrian Science Fund (FWF) Grants P21389-B03 and P24918-B21.
References
- Boyes, D. C., Zayed, A. M., Ascenzi, R., McCaskill, A. J., Hoffman, N. E., Davis, K. R. and Gorlach, J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13(7): 1499-1510.
- Borges, F., Gardner, R., Lopes, T., Calarco, J. P., Boavida, L. C., Slotkin, R. K., Martienssen, R. A. and Becker, J. D. (2012). FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods 8(1): 44.
- Honys, D. and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132(2): 640-652.
- Ibarra, C. A., Feng, X., Schoft, V. K., Hsieh, T. F., Uzawa, R., Rodrigues, J. A., Zemach, A., Chumak, N., Machlicova, A., Nishimura, T., Rojas, D., Fischer, R. L., Tamaru, H. and Zilberman, D. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337(6100): 1360-1364.
- Johnson-Brousseau, S. A. and McCormick, S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39(5): 761-775.
- Schoft, V. K., Chumak, N., Bindics, J., Slusarz, L., Twell, D., Kohler, C. and Tamaru, H. (2015). SYBR Green-activated sorting of Arabidopsis pollen nuclei based on different DNA/RNA content. Plant Reprod 28(1): 61-72.
- Schoft, V. K., Chumak, N., Choi, Y., Hannon, M., Garcia-Aguilar, M., Machlicova, A., Slusarz, L., Mosiolek, M., Park, J. S., Park, G. T., Fischer, R. L. and Tamaru, H. (2011). Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci U S A 108(19): 8042-8047.
- Schoft, V. K., Chumak, N., Mosiolek, M., Slusarz, L., Komnenovic, V., Brownfield, L., Twell, D., Kakutani, T. and Tamaru, H. (2009). Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep 10(9): 1015-1021.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chumak, N., Mosiolek, M. and Schoft, V. K. (2015). Sample Preparation and Fractionation of Arabidopsis thaliana Sperm and Vegetative Cell Nuclei by FACS. Bio-protocol 5(22): e1664. DOI: 10.21769/BioProtoc.1664.
Category
Plant Science > Plant cell biology > Organelle isolation
Cell Biology > Cell-based analysis > Flow cytometry
Cell Biology > Cell staining > Nucleic acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











