- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Substituted Cysteine Accessibility Method for Topology and Activity Studies of Membrane Enzymes Forming Thioester Acyl Intermediates in Bacteria
Published: Vol 5, Iss 21, Nov 5, 2015 DOI: 10.21769/BioProtoc.1640 Views: 8791
Reviewed by: Fanglian HeSoazig Le Guyon Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1769 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1584 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1927 Views
Abstract
The topology of membrane proteins and enzymes can be determined using various methods including reporter protein fusions and accessibility of cysteine residues to alkylating agents. Here we describe a variation of the substituted cysteine accessibility method to determine membrane topology and activity of enzymes containing an active site cysteine. Membrane topology of proteins can be predicted using different programs and the actual membrane topology can be determined by monitoring the accessibility of cysteine residues introduced in periplasmic (exposed) or cytoplasmic (not exposed) loops to alkylating agents. A two-step protocol is described where whole Escherichia coli (E. coli) cells are first treated with or without a membrane impermeable thiol reagent (2-sulfonatoethyl)-methane thiosulfonate (MTSES) and subsequently labeled with an alkylating reagent maleimide polyethyleneglycol (malPEG). When cysteine residues are accessible to MTSES, and thus exposed to (or accessible from) the periplasm, their free thiol groups covalently react with MTSES and consequently, are blocked for alkylation with malPEG. The thiol groups of cytoplasmic or membrane-embedded cysteine residues are not accessible to MTSES and proteins can be alkylated with malPEG resulting in an increase in molecular weight of 5 kDa. In the second part of the protocol, accessibility of cysteine residues is used to address the acylation state of enzymes that form stable thioester acyl intermediates. Thioesters can be specifically cleaved by neutral hydroxylamine, leading to a free thiol group of the active site cysteine that can then be alkylated with malPEG.
Materials and Reagents
- Escherichia coli K12 cells
- Distilled sterile water (dH2O)
- Acetone (Sigma-Aldrich, catalog number: 24201-1L-R )
- Chloroform (CHCl3) (VWR International, catalog number: 22711.290 )
- Trichloroacetic acid (TCA) (Sigma-Aldrich, catalog number: 27242-500G-R )
- Ethanol (EtOH) (100%) (Sigma-Aldrich, catalog number: 24103-5L-R )
Note: Product 24103 has been discontinued. - Tris(hydroxymethyl)aminomethane hydrochloride (Trizma base) (Sigma-Aldrich, catalog number: 337411KG )
- 1 M KH2PO4 (Merck Millipore Corporation, catalog number: A148973 )
- 1 M Potassium phosphate dibasic K2HPO4 (Fluka, catalog number: 60353 )
Note: Currently, it is “ Sigma-Aldrich, catalog number: 60353 ”. - Sodium dodecyl sulfate 20%SDS 20% (biosolve-chemicals, catalog number: 19812323 )
- Urea (Merck Millipore Corporation, Calbiochem®, catalog number: 66612 )
- NaOH (Merck Millipore Corporation, catalog number: 21778425 )
- Bromophenol blue (Sigma-Aldrich, catalog number: 114391 )
- Glycerol (VWR International, catalog number: 24388.295 )
- Parafilm (Thomas Scientific, Pichiney Plastic Packaging, catalog number: PM-992 )
- 50 mM Phosphate buffer (PB) (pH 7.0) (see Recipes)
- 250 mM Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich,catalog number: ED2SS ) (see Recipes)
- 0.3 M Sodium (2-sulfonatoethyl) methane thiosulfonate (MTSES) (Anatrace, catalog number: S110MT ) (see Recipes)
- 1 M L-cysteine (IGN, catalog number: 194646 ) (see Recipes)
Note: Currently, it is “mpbio, catalog number: 194646 ”. - Methanol (MeOH) (95%) (Sigma-Aldrich, catalog number: 32213-1L ) (see Recipes)
- 1 M Tris-HCl (see Recipes)
- Denaturation buffer (see Recipes)
- PEG buffer (see Recipes)
- 1.2 mM Methoxypolyethylene glycol 5000 maleimide (malPEG) (Sigma-Aldrich, catalog number: 63187 ) (see Recipes)
- Hydroxylamine solution (HA) (Sigma-Aldrich, catalog number: 59417 ) (see Recipes)
- 1 M Tris.HCl, 1% SDS (see Recipes)
- SDS-Sample buffer (see Recipes)
- DL-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D9779 ) (see Recipes)
Equipment
- Incubator (37 °C) (Infors AG)
- Spectrophotometer (600 nm) (Eppendorf, model: Biophotometer plus )
- Tabletop centrifuge (Eppendorf, model: 5472R/5427R )
- Gelectrophoresis apparatus (Bio-Rad Laboratories, model: Miniprotean II )
- Western blot apparatus (Bio-Rad Laboratories, model: Transblot Turbo )
- Vortex Genie 2 (Scientific Industries)
Procedure
I. Accessibility of cysteine residues using thiol specific and alkylating reagents
- Culture and induction
The culture conditions depend on the expression system and bacterial species used. For E. coli, IPTG inducible promoter such as pLac or L-arabinose inducible pBAD and systems alike are highly recommended. Use a single colony from a fresh plate streaked from -80 °C stock to inoculate a small volume (5 ml) for an overnight culture. Induce expression of the gene of interest in early exponential phase (OD600 around 0.2 in LB medium) and let bacteria grow at 37 °C till late exponential (2 h, OD600 around 0.6-0.7) to produce recombinant protein. This method has to be determined for the protein of interest. Measure absorbance at 600 nm (A600) at the end of induction period and chill culture on slushy ice. Slushy ice is 4 °C and cools the cell culture down quickly. The protocol is based on 1 ml of cells with an A600 of 1.0 (approximately 5 x 108-1 x 109 cells). - Preparation of samples for MTSES protection
Prior to experiment: Remove MTSES from -20 °C and let it warm up to room temperature, prepare fresh denaturation buffer and PEG buffer. All steps are performed at room temperature unless stated otherwise.- Centrifuge 2 x 1 ml of cell culture for 1 min. at 16,100 x g in tabletop centrifuge. Wash cells in 1 ml PB.
- Centrifuge cells. Resuspend cell pellet in 0.24 ml PB buffer and add 10 μl 25 mM EDTA (1 mM final). Use cells directly in step C.
Note: EDTA treatment permeabilizes the outer membrane. - For each sample, aliquot 120 μl into microcentrifuge tubes labeled with (+) and without (-) MTSES.
- Centrifuge 2 x 1 ml of cell culture for 1 min. at 16,100 x g in tabletop centrifuge. Wash cells in 1 ml PB.
- MTSES protection
Note: Treat samples as gently as possible while exposed to MTSES. EDTA permeabilizes the outer membrane, but the cytoplasmic membrane needs to be intact. After addition of L-cysteine to quench the reaction, samples can be treated more roughly. Wash steps are to remove L-cysteine.- Add 13 μl 0.3 M MTSES (30 mM final) to tube labeled (+) MTSES.
- Add 13 μl dH2O to tube labeled (-) MTSES by carefully pipetting up and down.
- Incubate 30 min at room temperature.
- Add 14 μl 1 M L-cysteine (100 mM final) to both tubes by carefully pipetting up and down.
- Incubate 10 min at room temperature.
- Add 0.75 ml PB.
- Centrifuge for 5 min at 16,100 x g in tabletop centrifuge.
- Wash the cells 2x with 0.5 ml PB.
- Resuspend cells in 50 μl PB.
- Add 13 μl 0.3 M MTSES (30 mM final) to tube labeled (+) MTSES.
- Sample denaturation with methanol/chloroform
Note: Pre-cool tabletop centrifuge to 4 °C.- Add 750 μl of Denaturation Buffer.
- Vortex vigorously, incubate at 4 °C for 30 min.
- Spin at 16,100 x g for 3 min.
- Remove supernatant very carefully and completely with pipette. An additional centrifugation step is usually not required.
- Add 400 μl 95% MeOH.
- Spin at 16,100 x g for 4 min.
- Remove supernatant carefully and air-dry pellets for 15 min at room temperature.
Note: Do not invert tubes, pellets are loose.
- Add 750 μl of Denaturation Buffer.
- Alkylation with malPEG
Note: Bring bottle with malPEG to room temperature and make malPEG solution, protect from light. Put malPEG powder back at -20 °C after wrapping bottle with Parafilm.- Cool 100% EtOH on ice.
- Resuspend each air-dried pellet in 100 μl PEG buffer, vortex.
- Add 20 μl malPEG to each tube and mix carefully by pipetting up and down.
- Incubate at room temperature for 30 min in the dark.
- Add 1.5 ml ice cold 100% EtOH and freeze at -20 °C for 1 h.
Note: Samples can be left at -20 °C overnight.
- Cool 100% EtOH on ice.
- SDS-PAGE and Western blotting
Note: Before starting, pre-cool tabletop centrifuge.- Pellet samples for 15 min at 16,100 x g in tabletop centrifuge.
- Carefully remove most of the supernatant.
- Re-centrifuge samples and remove all left over liquid.
- Air dry pellets.
Note: Do not invert tubes, pellets are loose. - Resuspend samples in SDS-PAGE sample buffer containing 4 mM DTT.
- Heat at 100 °C for 5 min.
- Load equivalent amounts on gel using A600 measurements at time of sample preparation.
- Load each -/+ pair adjacent to each other.
- Run gel and Western-blot.
- Pellet samples for 15 min at 16,100 x g in tabletop centrifuge.
II. Accessibility of cysteine to alkylating agents after cleavage of thioester bond
Follow protocol described in A.
- HA treatment
- Centrifuge 1.6 ml of cell culture for 1 min at 16,100 x g in tabletop centrifuge.
- Resuspend cells in 1 ml PB.
- Divide in 2 x 500 μl in tubes labeled – and + HA and centrifuge cells.
- Resuspend cells in 500 μl 1M Tris-HCl (pH 7.0), 1% SDS (-) or 500 μl 1 M HA (pH 7.4), 1% SDS (+) by pipetting up and down.
Note: This denatures proteins and cleaves thioester bonds in one step. - Incubate at room temperature for 30 min.
- Add 50 μl 100% TCA, vortex.
- Incubate on ice for 30 min. Pre-cool tabletop centrifuge.
- Centrifuge protein precipitate for 15 min at 16,100 x g at 4 °C.
- Wash pellet 3 x with 1 ml ice cold acetone (stored at -20 °C) and centrifuge 10 min at 16,100 x g each time.
- Air dry pellets. Follow steps in E and F.
Note: TCA pellets are often difficult to dissolve. Vortex rigorously and frequently.
- Centrifuge 1.6 ml of cell culture for 1 min at 16,100 x g in tabletop centrifuge.
Representative data
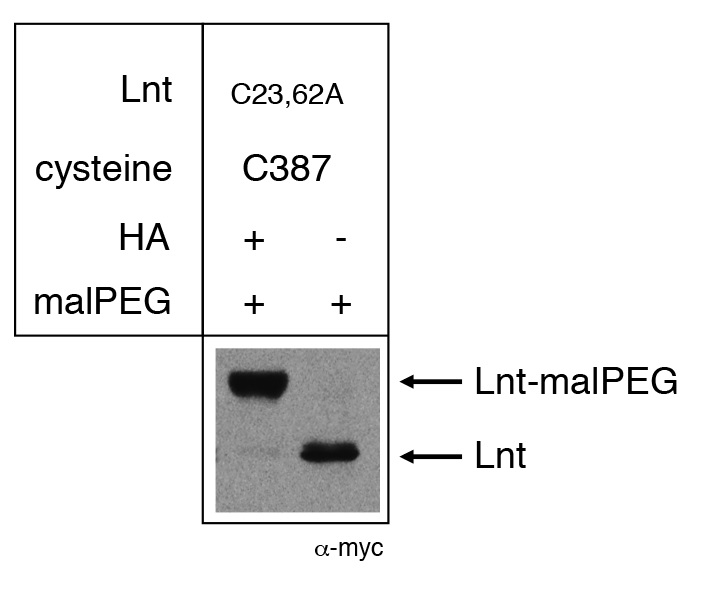
Figure 1. Example of malPEG alkylation profile of Apolipoprotein N-acyltransferase (Lnt) of E. coli. Cysteine residues 23 and 62 were substituted by alanine, leaving a single active site cysteine residue C387. Lnt forms a thioester acyl-enzyme intermediate in vivo with C387 and is blocked for reaction with malPEG. Upon treatment of cells with hydroxylamine (HA), the thioester bond is cleaved, leaving a free thiol group of C387 and alkylation by malPEG. Please see Gélis-Jeanvoine et al. (2015) for representative examples.
Notes
Results may vary depending on the membrane protein studied. It is highly recommended that negative and positive controls are included to validate technical details of the procedure. For example, use of empty vector, a cysteine-less variant of the protein of interest and inactive mutants with substituted cysteine residues are recommended.
Recipes
- Phosphate buffer (PB) (50 mM, pH 7.0)
Make 100 ml 1 M KH2PO4
Make 100 ml 1 M K2HPO4
Mix 1.5375 ml K2HPO4 with 0.9625 ml KH2PO4 and add distilled sterile water to 50 ml
The final concentration is 50 mM KPi (pH 7.0) - 0.25 M EDTA
Add 93.05 g EDTA to 800 ml of H2O
Adjust pH to 8.0 with NaOH
Note: Disodium EDTA will not go in solution until the pH around 8.0.
Divide into aliquots and autoclave - MTSES solution (make fresh before use)
For each 36.3 mg add 500 μl H2O, final concentration is 0.3 M - 1 M L-cysteine
121 mg L-cysteine in 1 ml H2O - 95% MeOH
To 47.5 ml MeOH add H2O to 50 ml - 1 M Tris.HCl
Add 121.1 g Trisma base into 800 ml H2O, adjust to pH 7.0 with HCl
Add H2O to final volume of 1 L
Autoclave - Denaturation buffer (make fresh before use)
To obtain a solution of H2O:MeOH:CHCl3 = 1:4:1, add to a 15 ml Falcon tube:
2 ml H2O
8 ml MeOH
2 ml CHCl3 - PEG buffer
Contains 10 M urea, 1% SDS, 1 mM EDTA in 1 M Tris pH 7.0.
Per ml solution:
0.6 g urea
0.01 g SDS
Dissolve in 600 μl 1 M Tris (pH 7.0), requires mixing and immersion in warm water
Do not put the solution on ice after preparation because urea will precipitate
Add 4 μl 0.25 M EDTA
Adjust volume to 1 ml with 1 M Tris (pH 7.0)
Note: PEG buffer contains urea, SDS and EDTA that denatures all proteins and renders all unprotected cysteine residues accessible to malPEG. - malPEG solution
Note: Make fresh before use, can not be stored.
Allow malPEG to get to room temperature
Weigh a small quantity, around 5 mg, on weigh paper
Add H2O to reach a final concentration of 1.2 mM MalPEG: Add 170 μl of H2O for 1 mg of malPEG
Note: For each sample 10 μl of 1.2 mM malPEG is needed, which corresponds to 12 nmol malPEG. 1 nmol malPEG is 5 μg. The minimum amount of malPEG to weigh accurately is 4-5 mg. This is enough for 34 samples. - Hydroxylamine solution (HA)
Dissolve 1.4 g HA in 10 ml of cold H2O to make 20 ml final of HA solution
Adjust the pH to 7.4 with 32% NaOH as follows
Add 1.5 ml of 32% NaOH, stir and check pH
Gradually add more NaOH and check pH till a pH of 7.4 is reached
Bring volume to 20 ml with H2O
Note: The pH should be between 6.9 and 7.5, around pH 8.5 solution loses specificity. - 1 M Tris.HCl, 1% SDS
Add 20% SDS to 1 M Tris solution to reach a final concentration of 1% - SDS-Sample buffer (4x)
2.5 ml 1 M Tris (pH 6.8)
1.0 g SDS
4 ml 100% glycerol
0.8 ml 0.1% bromophenol blue
0.16 ml 1 M DTT
Adjust total volume to 10 ml with dH2O - 1 M DTT
Dissolve 1.5 g DTT in 8 ml H2O, adjust volume to 10 ml
Divide in 1 ml aliquots and store at -20 °C
Acknowledgments
This work was financed by the Institut Pasteur.
References
- Bogdanov, M., Zhang, W., Xie, J. and Dowhan, W. (2005). Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): application to lipid-specific membrane protein topogenesis. Methods 36(2): 148-171.
- Gélis-Jeanvoine, S., Lory, S., Oberto, J. and Buddelmeijer, N. (2015). Residues located on membrane-embedded flexible loops are essential for the second step of the apolipoprotein N-acyltransferase reaction. Mol Microbiol 95(4): 692-705.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gélis-Jeanvoine, S. and Buddelmeijer, N. (2015). Substituted Cysteine Accessibility Method for Topology and Activity Studies of Membrane Enzymes Forming Thioester Acyl Intermediates in Bacteria. Bio-protocol 5(21): e1640. DOI: 10.21769/BioProtoc.1640.
Category
Microbiology > Microbial biochemistry > Protein > Structure
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









