- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of a Protein Exhibiting Isoleucine 2-epimerase Activity from Lactobacillus otakiensis JCM 15040
Published: Vol 5, Iss 20, Oct 20, 2015 DOI: 10.21769/BioProtoc.1632 Views: 8370
Reviewed by: Timo LehtiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Lauren E.-A. Eyssen [...] Raymond J. Owens
Mar 20, 2024 6247 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2097 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views
Abstract
Prominent accumulation of D-leucine, D-allo-isoleucine and D-valine was observed in the culture medium of the heterofermentative bacterial species, Lactobacillus otakiensis (L. otakiensis) JCM 15040. The racemase enzyme that resulted in this accumulation, isoleucine 2-epimerase, was purified from the bacterial cells. This is the first reported observation of such production of D-branched chain amino acids in lactic acid bacteria, and the first example of a racemase with isoleucine 2-epimerase activity in any organisms. In the described protocol, we introduce methods for purification of this protein from L. otakiensis JCM 15040. Because no specific ligand that has high affinity for this enzyme has been identified, the purification was performed using ammonium sulfate fraction, four types of column chromatography and preparative Native-PAGE, not using an affinity column chromatography. We hope that the protocol will provide useful information for purification of an enzyme that cannot easily be purified using an affinity column chromatography.
Keywords: Lactic acid bacteriaMaterials and Reagents
- 1.5 ml tube (ASONE Corporation, catalog number: 2-1998-02 )
- Dialysis membrane tube (molecular weight cutoff: 14,000) (EIDIA, catalog number: UC27-32-100 )
- Amicon Ultra 15 ml centrifugal filter 3 K device (Merck Millipore Corporation, catalog number: UFC500396 )
- Disposable homogenizer (1.5 ml scale) “Biomasher II” (NIPPI Corporation, catalog number: 320102 )
- Feeding tube (TERUMO CORPORATION, catalog number: SF-ET1725 )
- Syringe (10 ml scale) (TERUMO CORPORATION, catalog number: SS-10SZ )
- Plug silicon (ASONE Corporation, catalog number: 6-336-03 )
- Needle (TERUMO CORPORATION, catalog number: NN-2238R )
- Purification of the isoleucine 2-epimerase
- L. otakiensis JCM 15040 obtained from Japan Collection of Microorganisms (JCM)
- TOYOPEARL Phenyl-650M column
Note: Pack 50 ml of TOYOPEARL Phenyl-650M resin in a chromatography column (diameter: 2.5 cm, length: 10 cm) (Tosoh Bioscience LLC, catalog number: 14478 ). - TOYOPEARL Butyl-650M column
Note: Pack 50 ml of TOYOPEARL Butyl-650M resin in a chromatography column (diameter: 2.5 cm, length: 10 cm) (Tosoh Bioscience LLC, catalog number: 07477 ). - TOYOPEARL SuperQ-650M column
Note: Pack 50 ml of TOYOPEARL SuperQ-650M resin in a chromatography column (diameter: 2.5 cm, length: 10 cm) (Tosoh Corporation, catalog number: 17227 ). - Acrylamide (Wako Pure Chemical Industries, Siyaku, catalog number: 016-00765 )
- N, N’-Methylenebisacrylamide (Nacalai Tesque, catalog number: 22402-02 )
- Ammonium persulfate (Wako Pure Chemical Industries, Siyaku, catalog number: 7727-54-0 )
- N, N, N’, N’-Tetramethylethylenediamine (Nacalai Tesque, catalog number: 33401-72 )
- Ammonium sulfate (Wako Pure Chemical Industries, Siyaku, catalog number: 019-03435 )
- Sodium chloride (Wako Pure Chemical Industries, Siyaku, catalog number: 198-01675 )
- Lactobacilli MRS Broth (MRS medium powder) (BD, catalog number: 288130 ) (see Recipes)
- 50 mM sodium phosphate buffer (pH 7.2) containing 1 mM EDTA and 1 mM dithiothreitol (see Recipes)
Note: Unless otherwise indicated, this buffer is used as the standard buffer throughout the purification procedures.- Sodium dihydrogenphosphate dihydrate (Wako Pure Chemical Industries,
Siyaku, catalog number: 192-02815 ) - Disodium hydrogenphosphate 12-water (Wako Pure Chemical Industries,
Siyaku, catalog number: 196-02835 ) - Ethylenediamine-N, N, N', N'-tetraacetic acid, disodium salt, dehydrate (EDTA)
(Dojindo Molecular Technologies, catalog number: N001 ) - Dithiothreitol (Nacalai Tesque, catalog number: 14128-04 )
- Sodium dihydrogenphosphate dihydrate (Wako Pure Chemical Industries,
- Red Sepharose CL-4B column (Ohshima and Sakuraba, 1986) (see Recipes)
Note: According to Recipes, prepare Red Sepharose CL-4B resin by attaching Reactive Red 120 (Sigma-Aldrich, catalog number: R0378-50G ) to Sepharose® CL-4B (Sigma-Aldrich, catalog number: CL4B200-100ML ). Pack 10 ml of Red Sepharose CL-4B resin in a chromatography column (diameter: 1.5 cm, length: 12 cm). - 1.5 M Tris-HCl buffer (pH 8.8) (see Recipes)
- 0.5 M Tris-HCl buffer (pH 6.8) (see Recipes)
- 2-Amino-2-hydroxymethyl-1, 3-propanediol (Tris base)
- Hydrochloric acid
- 2-Amino-2-hydroxymethyl-1, 3-propanediol (Tris base)
- Native-PAGE electrophoresis buffer (see Recipes)
- 2-Amino-2-hydroxymethyl-1, 3-propanediol (Tris base)
- Glycine (Wako Pure Chemical Industries, Siyaku, catalog number: 077-00735 )
- 2-Amino-2-hydroxymethyl-1, 3-propanediol (Tris base)
- 2x Native-PAGE gel loading buffer (see Recipes)
- L. otakiensis JCM 15040 obtained from Japan Collection of Microorganisms (JCM)
- Isoleucine 2-epimerase activity assay
- Pyridoxal-5’-phosphate (Nacalai Tesque, catalog number: 29606-74 )
- L-isoleucine (PEPTIDE INSTITUTE, catalog number: 2712 )
- Flavin adenine dinucleotide disodium salt (Nacalai Tesque, catalog number: 16010-06 )
- 4-Aminoantipyrine (Nacalai Tesque, catalog number: 01907-52 )
- Phenol (Wako Pure Chemical Industries, Siyaku, catalog number: 168-12721 )
- D-Amino acid oxidase from porcine kidney (Sigma-Aldrich, catalog number: A5222-200UN )
- Peroxidase (TOYOBO BIO CHEMICAL DEPT, catalog number: PE0-301 )
- 0.5 M sodium phosphate buffer (pH 8.0) (see Recipes)
- Sodium dihydrogenphosphate dihydrate
- Disodium hydrogenphosphate 12-water
- Sodium dihydrogenphosphate dihydrate
- Pyridoxal-5’-phosphate (Nacalai Tesque, catalog number: 29606-74 )
Equipment
- Purification of the isoleucine 2-epimerase
- Incubator (TITEC CORPORATION, model: BR-43FM )
- Centrifuge (Hitachi Koki Co., model number: CR 21F and TOMY SEIKO CO., model number: MX-300 )
- Multi-Beads shocker (Yasui Kikai Corporation)
- Electrophoresis apparatus
- Column chromatography equipment (see Representative data)
- Magnetic stirrer (ASONE Corporation, model: HS-50E )
- Two-way stopcock (Bio-Rad Laboratories, catalog number: 7328102 )
- Three-way stopcock (TERUMO CORPORATION, catalog number: TS-TR1K )
- Chromatography column (diameter: 2.5 cm, length: 10 cm) (Bio-Rad Laboratories, catalog number: 7372512 )
- Chromatography column (diameter: 1.5 cm, length: 12 cm) (Bio-Rad Laboratories, catalog number: 7321010 )
- Fraction tube (15 ml scale) (ASONE Corporation, catalog number: 2-8007-02 )
- Incubator (TITEC CORPORATION, model: BR-43FM )
- Isoleucine 2-epimerase activity assay
- Spectrophotometer (Shimadzu Scientific Instruments, model number: UVmini-1240 )
- Water bath (TOKYO RIKAKIKAI CO., model number: NTT-20S )
- Vortex (TAITEC CORPORATION, catalog number: 0061271-000 )
- Spectrophotometer (Shimadzu Scientific Instruments, model number: UVmini-1240 )
Procedure
- Purification of the isoleucine 2-epimerase
- Prepare 5 test tubes including 10 ml of MRS medium for preculture.
- Inoculate a part of a colony of L. otakiensis JCM 15040 into each MRS medium (10 ml) in the test tube. The preculture is performed statically at 30 °C for 36 h.
- The main culture is started by addition of the preculture media (10 ml x 5 tubes = 50 ml) into 10 L of MRS medium.
- L. otakiensis JCM 15040 is cultivated statically at 30 °C for 30 h in an incubator, after which cells are pelleted by centrifugation (8,000 x g for 30 min at 4 °C). The cell pellet (ca. 32.2 g, wet weight) is used as the starting material for purification of protein exhibiting isoleucine 2-epimerase activity. Unless otherwise indicated, all purification procedures are carried out at room temperature, and the enzyme solution is stored at 4 °C.
- To prepare a crude extract, the cells are washed twice with about four volumes (120 ml) of the standard buffer, and suspend in 120 ml of the same buffer. On the occasion of the washing, the cells are collected by centrifugation (8,000 x g for 30 min at 4 °C). Next, they are disrupted using a Multi-Beads Shocker (2,500 rpm for 60 sec at 2 °C, 5 times), and centrifuged (10,000 x g for 30 min at 4 °C). The resultant supernatant (100 ml) is used as the crude extract.
- The crude extract (100 ml) is mixed with two volumes of 3.6 M (NH4)2SO4 dissolved in the standard buffer. After incubation on a magnetic stirrer for 4 h at 4 °C, the mixture is centrifuged (10,000 x g for 30 min at 4 °C), and the supernatant (280 ml) is retrieved.
- The collected supernatant (280 ml) is applied to a TOYOPEARL Phenyl-650M column. This column chromatography separates proteins on the basis of hydrophobic interactions between the proteins and the resin. Before the supernatant is loaded, pre-equilibrate the column with 10 column volumes (500 ml) of 2.4 M (NH4)2SO4 dissolved in the standard buffer. After the loading, the column is washed once with three column volumes (150 ml) of the same buffer and proteins are eluted using a linear gradient of 2.4 to 0.4 M (NH4)2SO4 in the buffer [used buffer: 250 ml of the buffer with 2.4 M (NH4)2SO4 and 250 ml of the buffer with 0.4 M (NH4)2SO4]. The elution is performed at a flow rate of about 1 ml/min, and 50 fractions including about 10 ml of elute are collected.
- Assay the enzyme activity in each fraction (see section B “Isoleucine 2-epimerase activity assay” below), and choose five active fractions showing higher activity than other fractions. The five active factions (about 50 ml) are mixed, and then dialyzed against 100 volumes (5 L) of the standard buffer at 4 °C. After 4 h, the standard buffer (5 L) is changed and dialysis is continuously performed at 4 °C for 12 h.
- This first dialysate (60 ml) is mixed with two volumes of 3.6 M (NH4)2SO4 dissolved in the standard buffer, and the mixture is applied to a TOYOPEARL Butyl-650M column. This column chromatography separates proteins on the basis of hydrophobic interactions between the proteins and the resin. Before the supernatant is loaded, pre-equilibrate the column with 10 column volumes (500 ml) of 2.4 M (NH4)2SO4 dissolved in the standard buffer. After the loading, the column is washed once with three column volumes (150 ml) of the same buffer, and proteins are eluted using a linear gradient of 2.4 to 0.4 M (NH4)2SO4 in the buffer [used buffer: 250 ml of the buffer with 2.4 M (NH4)2SO4 and 250 ml of the buffer with 0.4 M (NH4)2SO4]. The elution is performed at a flow rate of about 1 ml/min, and 50 fractions including about 10 ml of elute are collected. The five active fractions are pooled and dialyzed as described above.
- This second dialysate (60 ml) is applied to a TOYOPEARL SuperQ-650M column. This column chromatography separates proteins on the basis of ionic interactions between the proteins and the resin. Before the supernatant is loaded, pre-equilibrate the column with 10 column volumes (500 ml) of the standard buffer. After the loading, the column is washed once with three column volumes (150 ml) of the buffer, and proteins are eluted using a linear gradient of 0 to 250 mM NaCl in the buffer (used buffer: 250 ml of the standard buffer and 250 ml of the buffer with 250 mM NaCl). The elution is performed at a flow rate of about 1 ml/min, and 50 fractions including about 10 ml of elute are collected. The five active fractions are pooled and dialyzed as described above.
- SDS-PAGE of this third dialysate shows that this dialysate includes not only isoleucine 2-epimerase, but also a putative NAD+-dependent alcohol dehydrogenase (data not shown). To remove this NAD+-dependent dehydrogenase, the third dialysate (50 ml) is applied to a Red Sepharose CL-4B column. Because the red dye immobilized in this column binds to a wide variety of NAD+- and NADP+-dependent enzymes but not isoleucine 2-epimerase, the NAD+-dependent enzyme and isoleucine 2-epimerase are separated by pooling the flow-through of this column chromatography. Before the supernatant is loaded, pre-equilibrate the column with 10 column volumes (100 ml) of the standard buffer. After the loading, the column is washed the column once with three column volumes (30 ml) of the standard buffer. During sample loading and column washing, the flow-through (about 80 ml) as the active fractions is pooled. The resultant enzyme solution is then concentrated to 300 μl using an Amicon Ultra centrifugal filter 3 K device, and then the concentrated enzyme solution is stored at 4 °C.
- Native-polyacrylamide gel electrophoresis (Native-PAGE) of the concentrated enzyme solution (300 μl) is performed on a polyacrylamide slab gel for further enzyme purification; 30 μl of the enzyme solution is loaded on each of 10 lanes. The electrophoresis is performed using the method of Laemmli (Laemmli, 1970) with some modifications; buffers without sodium dodecyl sulfate are used, and the protein sample is not heated during the pretreatment procedures. After electrophoresis using a constant current of 20 mA for 90 min, the gel is cut into 16 pieces using a cutter and a ruler. The gel pieces are individually crushed in 300 μl of the standard buffer using Biomasher II, and the resultant solutions are centrifuged (17,000 x g for 15 min at 4 °C) (Figure 2). The enzyme activity of the supernatants is assayed and the active enzyme solution is used as the final purified enzyme solution from L. otakiensis JCM 15040.
- Prepare 5 test tubes including 10 ml of MRS medium for preculture.
- Isoleucine 2-epimerase activity assay
- Prepare the first step reaction mixture in 1.5 ml tube. The composition of the first step reaction mixture is shown in the following table. Run this reaction for 1 h at 30 °C. The 1.5 ml tube containing the reaction mixture is incubated in a water bath.
The first step reaction mixture (500 μl) 0.5 M sodium phosphate buffer (pH 8.0) 100 μl 0.5 mM pyridoxal-5’-phosphate 100 μl 50 mM L-isoleucine 100 μl Enzyme solution 50 μl H2O 150 μl - Boil the first step reaction mixture in 1.5 ml tube for 10 min, and thus cool it to room temperature.
- Prepare the second step reaction mixture by adding reagents shown in the following table into the first step reaction mixture. Run this reaction for 15 min at 37 °C. The 1.5 ml tube containing the reaction mixture is incubated in a water bath.
The second step reaction mixture (1,000 μl) The first step reaction mixture 500 μl 0.5 M sodium phosphate buffer (pH 8.0) 100 μl 0.4 mM fravin-adenine dinucleotide 50 μl 4 mM 4-aminoantipyrine 50 μl 40 mM phenol 50 μl 5 Unit/ml D-amino acid oxidase 50 μl 20 Unit/ml horseradish peroxidase 50 μl H2O 50 μl - Apply the second step reaction mixture to a spectrophotometer, and thus measure absorbance at 500 nm. A measurement of the reaction mixture without the enzyme solution is used as a blank data.
- Prepare the first step reaction mixture in 1.5 ml tube. The composition of the first step reaction mixture is shown in the following table. Run this reaction for 1 h at 30 °C. The 1.5 ml tube containing the reaction mixture is incubated in a water bath.
Representative data
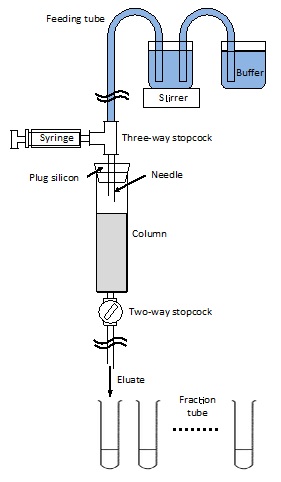
Figure 1. Image of the column chromatography equipment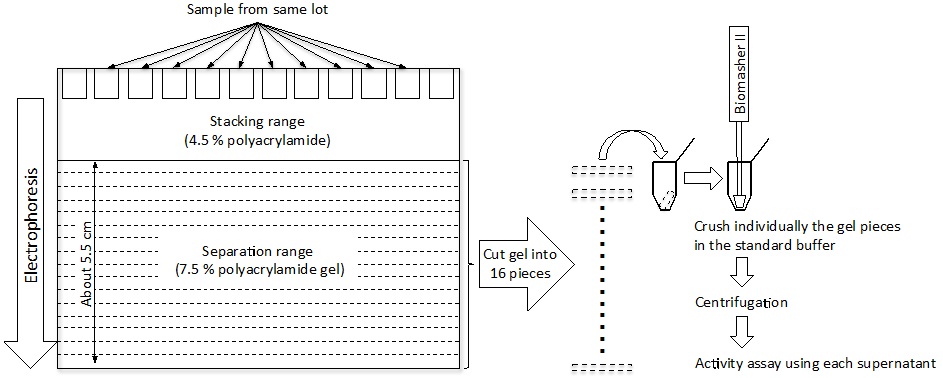
Figure 2. Image of the purification procedure using Native-PAGE
Notes
Isoleucine 2-epimerase activity assay
For choosing active fraction, the enzyme activity is rapidly assayed by the following 2 step reactions (Figure 3). At first step, isoleucine 2-epimerase reaction with L-isoleucine as a substrate produces D-allo-isoleucine. At next step, D-amino acid oxidase-peroxidase coupling reaction is performed. In this coupling reaction, D-amino acid oxidase reaction produces H2O2 in oxidization of D-allo-isoleucine, and thus peroxidase reaction produces indophenol compound (λmax: 500 nm, ε: 6.39 mM-1.cm-1) from 4-aminoantipyrine and phenol using oxidation power of H2O2. The isoleucine 2-epimerase activity is indirectly assayed by increase in absorbance at 500 nm occurring from the indophenol compound production. 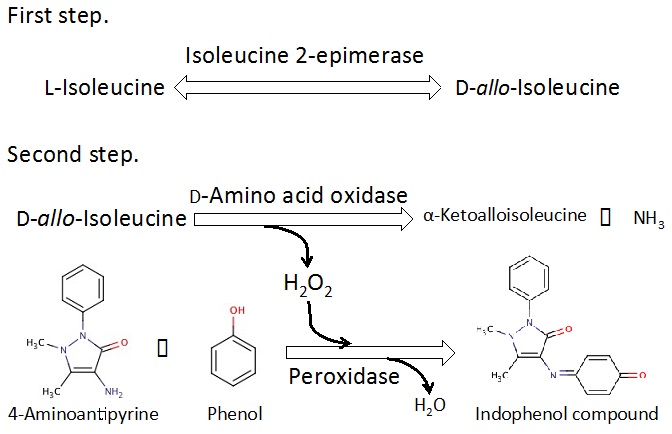
Figure 3. Principle of the assay method for isoleucine 2-epimerase activity
Recipes
- Purification of the isoleucine 2-epimerase
- MRS medium (1 L)
Mix 55 g of MRS medium powder with 800 ml of dH2O
Add dH2O to 1,000 ml
Autoclave
Stored at room temperature - 50 mM sodium phosphate buffer (pH 7.2) containing 1 mM EDTA and 1 mM dithiothreitol (1 L)
Mix 3.90 g of NaH2PO4.2H2O and 8.95 g of Na2HPO4.12H2O with 800 ml of dH2O
Add 2 ml of 0.5 M EDTA (pH 8.0)
pH to 7.2 with HCl or NaOH (aq)
Add dH2O to 1,000 ml
Autoclave
Add and mix 0.154 g of dithiothreitol
Stored at 4 °C - Red Sepharose CL-4B
Add Reactive Red 120 (1 g in 100 ml of dH2O) and NaCl (22%, 20 ml) into slurry of Sepharose 4B (100 ml) previously washed with dH2O
Tumble the mixture at 25 °C for 30 min slowly
Add 2 g of Na2CO3 to the mixture
Coupling is achieved by shaking gently for about 4 days at 25 °C
Wash the slurry with 2 L of dH2O and 2 L of 1 M KCl
Store the Red-Sepharose 4B at 4 °C - 1.5 M Tris-HCl buffer (pH 8.8, 100 ml)
Mix 18.2 g of Tris base with 80 ml of dH2O
pH to 8.8 with HCl
Add dH2O to 100 ml
Autoclave
Store at room temperature - 0.5 M Tris-HCl buffer (pH 6.8, 100ml)
Mix 6.1 g of Tris base with 80 ml of dH2O
pH to 6.8 with HCl
Add dH2O to 100 ml
Autoclave
Stored at room temperature - Polyacrylamide slab gel for Native-PAGE (thickness, 1 mm; wide, 106 mm; high, 80 mm)
Separate range (7.5% polyacrylamide gel, 6 ml) 1.5 M Tris-HCl buffer (pH 8.8) 1.5 ml 29% (w/v) acrylamide + 1% (w/v) N, N’-methylene bis acrylamide solution 1.5 ml 10% (w/v) ammonium persulfate 200 μl dH2O 2.8 ml N, N, N’, N’-Tetramethylethylenediamine 3 μl Stacking range (4.5% polyacrylamide gel, 3 ml) 0.5 M Tris-HCl buffer (pH 6.8) 0.75 ml 29% (w/v) Acrylamide + 1% (w/v) N’, N’-methylene bis acrylamide solution 0.45 ml 10% (w/v) Ammonium peroxodisulphate 100 μl dH2O 1.7 ml N, N, N’, N’-Tetramethylethylenediamine 3 μl - Native-PAGE electrophoresis buffer (1 L)
Mix 3.03 g of Tris base, 14.4 g of glycine and 900 ml of dH2O
Add dH2O to 1 L
Autoclave
Stored at room temperature - 2x Native-PAGE gel loading buffer (10 ml)
Mix 2.5 ml of 0.5 M Tris-HCl buffer (pH 6.8), 1 ml of 2-mercaptoethanol, 1 g of sucrose, 1 mg of bromophenol blue and 8 ml of dH2O
Add dH2O to 10 ml
Stored at −20 °C
- MRS medium (1 L)
- Isoleucine 2-epimerase activity assay
- 0.5 M sodium phosphate buffer (pH 8.0, 100 ml)
Mix 1.06 g of NaH2PO4.2H2O and 15.5 g of Na2HPO4.12H2O with 80 ml of dH2O
pH to 8.0 with HCl or NaOH (aq)
Add dH2O to 100 ml
Autoclave
Stored at room temperature
- 0.5 M sodium phosphate buffer (pH 8.0, 100 ml)
Acknowledgments
In this method, we have modified Laemmli’s method for Native-PAGE. In addition, this work was supported by a grant for Promotion of Basic Research Activities for Innovate Bioscience from the Bio-oriented Technology Research Advancement Institution (BRAIN) and JSPS KAKENHI Grant Number 2402734.
References
- Ohshima, T. and Sakuraba, H. (1986). Purification and characterization of malate dehydrogenase from the phototrophic bacterium, Rhodopseudomonas capsulata. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology 869(2): 171-177.
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mutaguchi, Y. and Ohshima, T. (2015). Purification of a Protein Exhibiting Isoleucine 2-epimerase Activity from Lactobacillus otakiensis JCM 15040. Bio-protocol 5(20): e1632. DOI: 10.21769/BioProtoc.1632.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









