- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bacterial Porphyrin Extraction and Quantification by LC/MS/MS Analysis
Published: Vol 5, Iss 19, Oct 5, 2015 DOI: 10.21769/BioProtoc.1616 Views: 9782
Reviewed by: Valentine V TrotterAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1775 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1825 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1790 Views
Abstract
Heme is an iron-containing porphyrin which acts as a prosthetic group in several enzymes involved in disparate functions, such as respiration and H2O2-scavenging. Escherichia coli is able to produce heme endogenously since it contains all the enzymes involved in the nine-step biosynthesis pathway, which in absence of stress and in iron-replete media proceeds unabated. However, we recently showed that two steps are affected by H2O2 stress (Mancini and Imlay, 2015). To compensate, two enzymes, namely the ferrochelatase (HemH) and an isozyme of coproporphyrinogen III oxidase (HemF), are activated by the H2O2-responsive regulator OxyR. Genetic mutations that block either adaptation cause the intracellular accumulation of protoporphyrin IX and coproporphyrinogen III, the substrates of HemH and HemF, respectively. We here describe a method used to extract and quantify protoporphyrin IX and coproporphyrin III, the product of the spontaneous oxidation of coproporphyrinogen III.
Keywords: HemeMaterials and Reagents
- Bacterial cell culture
- Ethyl acetate (Sigma-Aldrich, catalog number: 34972-1 L-R )
- Acetic acid (Sigma-Aldrich, catalog number: 45754-100 ML-F )
- Hydrochloric acid (Sigma-Aldrich, catalog number: H1758-100 ML )
- Protoporphyrin IX (Frontier Scientific, catalog number: P562-9 )
- Coproporphyrin III dihydrochloride (Frontier Scientific, catalog number: C-654-3 )
- Formic acid (Sigma-Aldrich, catalog number: 14265-1ML )
- Acetonitrile (Sigma-Aldrich, catalog number: 34967-250 ML )
Equipment
For porphyrin extraction
- Sonicator (Fisher Scientific, model: 550 sonic dismembrator )
For the LC/MS/MS analysis
- 5500 QTRAP LC/MS/MS system (AB Sciex)
- 1200 series HPLC system (Agilent Technologies)
- Degasser Autosampler (Agilent Technologies)
- Binary pump (Agilent Technologies)
- Zorbax SB-Aq column (4.6 x 50 mm, 5 µm) (Agilent Technologies, catalog number: 846975 )
Procedure
For the porphyrin extraction (adapted from Nakayashiki and Inokuchi, 1997)
- Culture bacterial cells until a final cell suspension equivalent to 100 ml with an A600 of 0.4.
- Harvest the cells by centrifugation at 7,000 x g at 4 °C for 10 min and wash the pellet in 20 ml pre-chilled 0.05 M Tris pH 8.2-2 mM EDTA.
- Resuspend the cells in 10 ml of the same buffer.
- Measure the A600 of the culture suspension and adjust to equalize the A600 of each sample by adding Tris-EDTA buffer.
- Spin down cells at 7,000 x g at 4 °C for 10 min. Resuspend the cell pellets in 1 ml ethyl acetate/acetic acid (3:1, v/v).
- Lyse the cells by sonication for 2 min with power 3 on ice.
- Remove the cell debris by centrifugation at 7,000 x g at 4 °C for 10 min and then transfer supernatant to a fresh tube.
- Add 1 ml H2O, vortex, and centrifuge at 7,000 x g for 5 min. Remove and discard the aqueous top layer.
- Repeat step 8.
- Add 100 µl of 3 M HCl to solubilize the porphyrins.
- Centrifuge after vigorous vortexing before transferring upper layer containing the water-soluble porphyrins into a fresh Eppendorf tube.
For the LC/MS/MS analysis
- Perform the LC separation with Zorbax SB-Aq column.
- Use 0.1% formic acid in water as mobile phase A and 0.1% formic acid in acetonitrile as mobile phase B.
- Use a flow rate of 0.3 ml/min.
- Perform a stepwise elution as follows: 0-1 min 100% A, then gradually decrease down to 5% A until min 10. Maintain 5% A until min 18 and then increase to 100% A until min 19. Maintain 100% A until the end (min 24).
- Set the autosampler at 5 °C.
- Inject sample (1 μl) from step 11 above.
- Use 0.1% formic acid in water as mobile phase A and 0.1% formic acid in acetonitrile as mobile phase B.
- Acquire the mass spectra with positive electrospray ionization (ESI) with the ion spray voltage set at 5,500 V.
- Set the source temperature at 450 °C.
- Set the curtain gas, ion source gas 1, and ion source gas 2 at 32, 65, and 50, respectively.
- Use multiple reactions monitoring (MRM) to monitor coproporphyrin III (m/z 655.4 --> m/z 596.3) and protoporphyrin IX (m/z 563.2 --> m/z 504.1). Use 1 µg of the porphyrin standards for the initial tests.
- Set the source temperature at 450 °C.
- Measure the peak areas of the resulting chromatograms with Analyst 1.6.
Note that this is a qualitative assessment. A standard curve using the corresponding porphyrin references is required to obtain absolute values.
Representative data
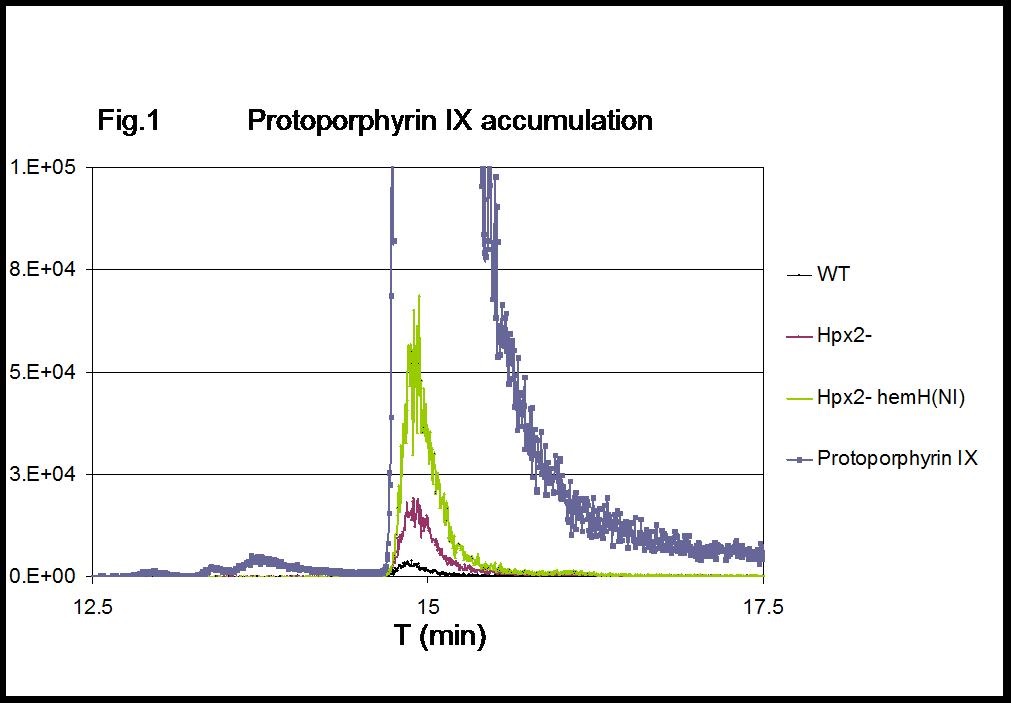
Figure 1. MRM chromatograms of protoporphyrin IX. Intracellular accumulation of protoporphyrin IX in H2O2-stressed cells (Hpx2-) is exacerbated by the lack of hemH activation [Hpx2- hemH(NI)]. 1 µg protoporphyrin IX was included as reference standard.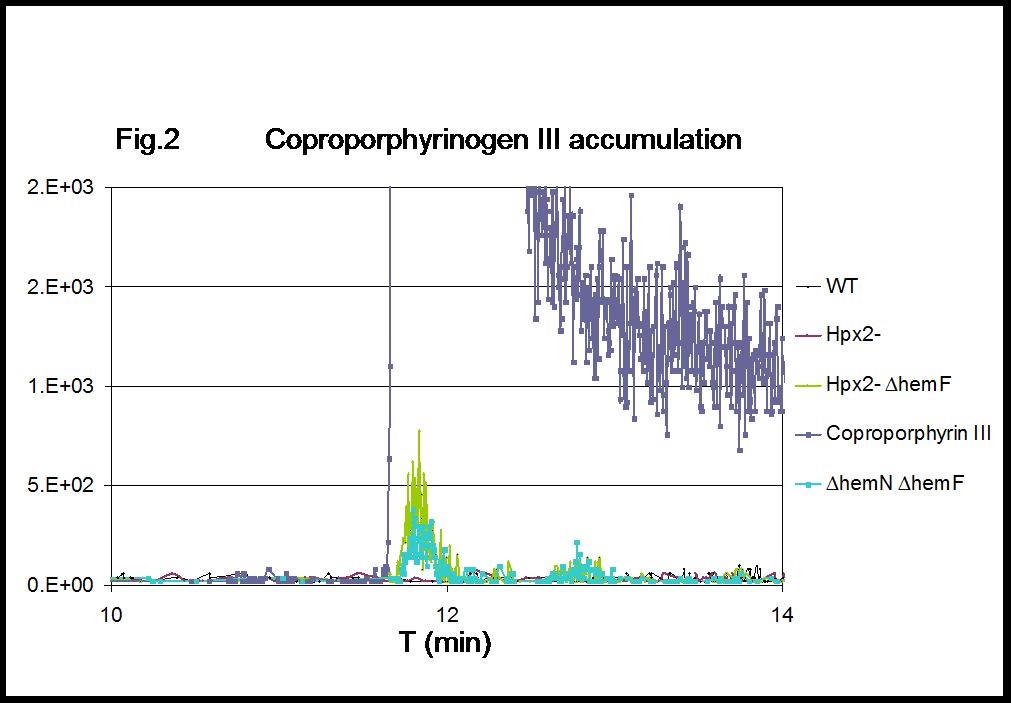
Figure 2. MRM chromatograms of coproporphyrin III. Intracellular coproporphyrinogen III accumulates in H2O2-stressed cells lacking hemF (Hpx2- ∆hemF), with levels comparable to those detected in cells lacking both the coproporphyrinogen III oxidase isozymes (∆hemN ∆hemF). 1 µg coproporphyrin III was included as reference standard.
Notes
Upon exposure to air all porphyrinogens autoxidize to porphyrins. Therefore, coproporphyrin III and not coproporphyrinogen III (the actual substrate of the two coproporphyrinogen III oxidase isozymes HemN and HemF) can be analyzed and quantified by LC/MS/MS. Similarly, protoporphyrinogen IX, the substrate of the penultimate enzyme of the heme biosynthesis pathway, namely protoporphyrinogen IX oxidase, spontaneously oxidizes to protoporphyrin IX. Therefore, protoporphyrin IX levels should reflect the sum of protoporphyrinogen IX and protoporphyrin IX.
Acknowledgments
This work was supported by grant GM49640 from the National Institutes of Health and by award number PBBEP3_139397 from the Swiss National Science Foundation. The protocol for the porphyrin extraction was adapted from Nakayashiki and Inokuchi (1997).
References
- Mancini, S. and Imlay, J. A. (2015). The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol Microbiol 96(4): 744-763.
- Nakayashiki, T. and Inokuchi, H. (1997). Effects of starvation for heme on the synthesis of porphyrins in Escherichia coli. Mol Gen Genet 255(4): 376-381.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mancini, S. and Imlay, J. A. (2015). Bacterial Porphyrin Extraction and Quantification by LC/MS/MS Analysis. Bio-protocol 5(19): e1616. DOI: 10.21769/BioProtoc.1616.
Category
Microbiology > Microbial biochemistry > Other compound
Biochemistry > Other compound > Porphyrins
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









