- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
TCRβ Clonotype Analysis of EBV and CMV-specific Human CD8+ T Cells
Published: Vol 5, Iss 19, Oct 5, 2015 DOI: 10.21769/BioProtoc.1606 Views: 8734
Reviewed by: Kathrin SutterMarielle CavroisAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Virus Infection and Titration of SARS-CoV in Mouse Lung
Craig Fett [...] Stanley Perlman
Mar 20, 2014 14183 Views

Multicolor Stimulated Emission Depletion (STED) Microscopy to Generate High-resolution Images of Respiratory Syncytial Virus Particles and Infected Cells
Masfique Mehedi [...] Ursula J. Buchholz
Sep 5, 2017 10124 Views

A Triple-challenge Mouse Model of Allergic Airway Disease, Primary Influenza Infection, and Secondary Bacterial Infection
Sean Roberts [...] Yoichi Furuya
Apr 20, 2020 4533 Views
Abstract
This protocol describes the quantification of all expressed T-cell antigen receptor (TCR) gene products within sorted (by flow cytometry) EBV and CMV-specific memory CD8+ T-cell populations using a template-switch anchored reverse transcription polymerase chain reaction (RT-PCR).
Materials and Reagents
- Nuclease-free 1.5 ml tubes (Sarstedt, catalog number: 72692005 )
- RNAlater (Life Technologies, catalog number: AM7020 )
Note: Currently, it is “Thermo Fisher Scientific, AmbionTM, catalog number: AM7020”. - FACS-sorted CD8+ T cells in RNAlater (Koning et al., 2013)
- T-cell markers CD3 (PerCP) (BD), CD8 (AmCyan/V500) (BD) and pMHC-tetramer (APC/PE) (home-made), and the dump markers CD14 and CD19 (both BioLegend, Pacific Blue®)
- LIVE/DEAD Viability dye (Thermo Fisher Scientific, InvitrogenTM)
- µMACS mRNA isolation kit (Miltenyi Biotec, catalog number: 130-075-201 )
- SMARTer Pico PCR cDNA synthesis kit (Takara Bio Company, Clontech, catalog number: 634928 )
- dNTPs 25 mM each (Life Technologies, catalog number: R1121 )
- Recombinant RNasin Ribonuclease Inhibitor 20 U/μl (Promega Corporation, catalog number: N2511 )
- Superscript II RNase H- Reverse transcriptase 200 U/μl (Thermo Fisher Scientific, InvitrogenTM, catalog number: 18064014 )
Note: This kit also contains dithiothreitol (DTT, 100 mM). - Nucleospin Extract II, gel and PCR clean up (MACHEREY-NAGEL GmbH & Co, catalog number: MN740609.250 )
- Molecular grade H2O (Sigma-Aldrich, catalog number: W4502-1L )
- Advantage 2 Polymerase Mix (Takara Bio Company, Clontech, catalog number: 639201 )
- Molecular grade agarose (Thermo Fisher Scientific, Fisher BioReagents®, catalog number: 10688973 )
- SYBR® Gold Nucleic Acid Gel Stain (Life Technologies, catalog number: S-11494 )
Note: Currently, it is “Thermo Fisher Scientific, InvitrogenTM, catalog number: S-11494”. - 1 kb Plus DNA ladder (Life Technologies, catalog number: 10787-018 )
Note: Currently, it is “Thermo Fisher Scientific, InvitrogenTM, catalog number: 10787-018 ”. - pGEM-T Easy Vector System kit I (Promega Corporation, catalog number: A1360 )
- Chemically competent E. coli DH5α (Home-made)
- AmpliTaq DNA Polymerase with buffer II (Life Technologies, catalog number: N8080153 )
Note: Currently, it is “Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: N8080153”. - Big Dye Terminator v3.1 cycle kit (Life Technologies, catalog number: 4337455 )
Note: Currently, it is “Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4337455”. - RNase-inactivator (RNase Away) (MP Biomedicals)
- Primers (can be ordered from any company that provides custom made DNA oligo’s):
*The final GGG bases in this oligo are RNA basesSMART II oligo 5’-AAGCAGTGGTATCAACGCAGAGTACGCGGG*-3’ Universal
Primer Long
5’-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3’Universal
Primer Short
5’-CTAATACGACTCACTATAGGGC-3’MBC2 5’-TGCTTCTGATGGCTCAAACACAGCGACCT-3’ M13 Forward 5’-TTTTCCCAGTCACGAC-3’ M13 Reverse 5’-CAGGAAACAGCTATGAC-3’ Oligo d(T)25 5’-d(T)25VN**-3’
**V = A, C, or G. N = A, C, G, or T
Note: Preferably, order primers with high purity (HPLC purified). - 10x Universal Primer mix (see Recipes)
- 50 mM DTT (see Recipes)
Equipment
- FACSAria III flow cytometer (BD)
- Heating blocks (42 °C and 70 °C) (VWR International)
- T-M-AR, DNA/RNA UV-cleaner box (Laminar flow hood/ UVC) (Biosan, catalog number: BS-040104-A06 )
- Standard table top Eppendorf centrifuge (for use at room temperature and 4 °C)
- PCR thermo cycler
Procedure
- FACS sort of antigen-specific T cells
- In the studies conducting this protocol, EBV and CMV-specific memory CD8+ T-cell populations were sorted using pMHC class I tetramers on a FACSAria III flow cytometer. A high fidelity staining strategy was used, comprising the specific T-cell markers CD3, CD8 and pMHC-tetramer, and the dump markers CD14 and CD19 to exclude any contamination during the sort from monocytes and B cells, respectively. In addition, a LIVE/DEAD Viability dye was included in the cell staining. For details on the specific staining, see Koning et al. (2013). Tetramer-positive populations usually comprised 0.1-1.0% of CD8+ T cells and in general 500-5,000 antigen-specific cells were isolated.
Note: Although the specific fluorochromes noted here were used in Koning et al. (2013), we have performed a similar staining strategy using different fluorochromes in other studies. Similarly, although this protocol specifies the analysis of EBV and CMV-specific T cells, there are no restrictions for the analysis of T cells directed against other species/antigens.
To obtain a complete and accurate view of the TCR composition of an antigen-specific T-cell population, make sure to sort a sufficient number of cells (at least 500-1,000 cells, ideally 5,000 cells or even more) at high purity (by including dump markers and a viability dye, as described above). Sort cells directly in RNAlater and store at -80 °C.
- In the studies conducting this protocol, EBV and CMV-specific memory CD8+ T-cell populations were sorted using pMHC class I tetramers on a FACSAria III flow cytometer. A high fidelity staining strategy was used, comprising the specific T-cell markers CD3, CD8 and pMHC-tetramer, and the dump markers CD14 and CD19 to exclude any contamination during the sort from monocytes and B cells, respectively. In addition, a LIVE/DEAD Viability dye was included in the cell staining. For details on the specific staining, see Koning et al. (2013). Tetramer-positive populations usually comprised 0.1-1.0% of CD8+ T cells and in general 500-5,000 antigen-specific cells were isolated.
- mRNA isolation
- Isolate mRNA using the µMACS mRNA Isolation kit, according to the manufacturer’s instructions. Elute with 30 µl preheated elution buffer.
- Store mRNA at -80 °C or proceed with cDNA synthesis.
- Isolate mRNA using the µMACS mRNA Isolation kit, according to the manufacturer’s instructions. Elute with 30 µl preheated elution buffer.
- cDNA generation
- Incubate 5.5 µl mRNA and 0.5 µl Oligo d(T)25 (25 mM) at 2 min at 70 °C followed by 2 min at 42 °C.
Note: Proceed as fast as possible after this step. Time can be won by preparing the mastermix below during the initial incubation. Ideally, preparation of the mastermix is finished at the same time as the incubation. - Add:
2.2 µl 5x First Strand buffer from SMARTer cDNA synthesis kit 0.4 µl DTT (50 mM) 0.4 µl dNTPs (25 mM) 1 µl SMART II Oligo (12 µM) from SMARTer cDNA synthesis kit or custom made* 0.5 µl RNasin (20 U/µl) 1 µl SuperScript II Reverse Transcriptase (200 U/µl) - Incubate 2 h at 42 °C.
Note: *Whilst a custom made SMART II oligo can be cost-reducing, it may perform less than the oligo that is included in the SMARTer cDNA synthesis kit.
- Incubate 5.5 µl mRNA and 0.5 µl Oligo d(T)25 (25 mM) at 2 min at 70 °C followed by 2 min at 42 °C.
- Purification of cDNA
Purify the cDNA using the Nucleospin Extract II Gel and PCR Clean Up kit. Follow the manufacturer’s protocol, with the exceptions described below.
Add to the cDNA:
77 µl H2O
13 µl buffer NTI
(Total volume: 100 µl)
A single wash step with 650 µl buffer NT3 suffices. Elute in 25 µl buffer NE.
Preferably, continue directly to the RACE (rapid amplification of cDNA ends) TCRβ PCR or store the purified cDNA at -80 °C. - RACE TCRβ-PCR
RACE PCR is performed using TCR-specific and anchor-complementary primers.
Prepare a PCR mastermix of all reagents except for the template. Prepare enough for a duplicate of each sample and one negative control (water instead of template). Small, artificial variations in clonotype composition can be caused by the PCR. Therefore, when a very accurate examination of the T-cell repertoire is desired (for example, in a longitudinal analysis), it is advised to perform a duplicate PCR that can be pooled after agarose gel electrophoresis (step 5).Per sample 5 µl 10x Advantage 2 SA PCR buffer 10 µl 10x Universal Primer mix (forward primers) 1 µl MBC2 (25 µM, reverse primer) 1 µl dNTPs (10 mM) 1 µl 50x Advantage 2 Polymerase mix 12 µl Purified cDNA 20 µl H2O 50 µl Total volume - Run reactions in a thermocycler under the following conditions:
Store the TCRβ products at -20 °C.1 cycle 95 °C 30 sec 5 cycles: 95 °C 15 sec 72 °C 2 min 5 cycles: 95 °C 15 sec 70 °C 30 sec 72 °C 2 min 30 cycles: 95 °C 15 sec 68 °C 30 sec 72 °C 2 min 4 °C ∞
Note: Do not store at 4 °C, as the A overhang of the PCR products will degrade over time.
- Isolation of Vβ products
- Run an electrophoresis on a 1% agarose gel, using a 1 kb DNA ladder to determine the size of the fragments. Leave an empty well between each sample to minimize the chance of cross-contamination. A band at 500-600 bp should be visible. Carefully excise the band of interest and transfer the gel to a clean 1.5 ml microcentrifuge tube.
- Extract the amplified TCR products from the gel using the NucleoSpin Extract II Gel and PCR Clean Up kit, according to the manufacturer’s instructions. Store samples at -20 °C.
Note: Perform the extra wash steps described in the protocol and also use preheated (37 °C) wash buffer NT3. Elute in 20 µl buffer NE.
- Run an electrophoresis on a 1% agarose gel, using a 1 kb DNA ladder to determine the size of the fragments. Leave an empty well between each sample to minimize the chance of cross-contamination. A band at 500-600 bp should be visible. Carefully excise the band of interest and transfer the gel to a clean 1.5 ml microcentrifuge tube.
- Ligation of Vβ product into plasmid and sequencing
Amplified TCR transcripts are ligated into the pGEM-T Easy vector. Prepare 15 µl ligation reactions, using 5 µl of sample, 7.5 µl Rapid ligation buffer, 1 µl pGEM-T Easy Vector, and 1.5 µl T4 DNA ligase. Incubate overnight (16-24 h) at 4 °C, then transform into chemically competent E. coli DH5α bacteria using 7.5 µl of the ligation mixture. Pick 96 single transformed bacterial clones to amplify using M13 primers. Use the following PCR mix per sample (=per bacterial colony):Use the following cycling conditions:2.5 µl AmpliTaq PCR Buffer II 0.2 µl M13 Forward primer (25 µM) 0.2 µl M13 Reverse primer (25 µM) 0.5 µl dNTPs (10 mM) 0.125 µl AmpliTaq DNA polymerase 3.0 µl MgCl2 (25 mM) 18.475 µl H2O 25 µl Total volume
Assess the efficiency of amplification on a 1% agarose gel. Sanger sequencing is performed using the Big Dye Terminator v3.1 Cycle kit and samples are sent to a high throughput facility to be sequenced by capillary electrophoresis.1 cycle 95 °C 30 sec 40 cycles: 95 °C 15 sec 57 °C 30 sec 72 °C 90 sec Hold: 4 °C ∞
Representative data
Example of a TCRβ-PCR: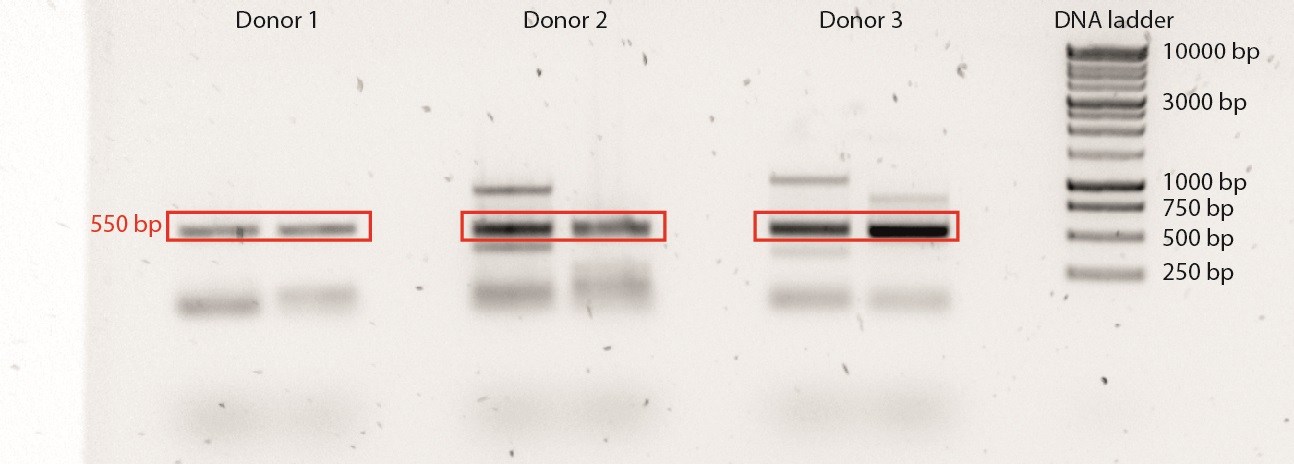
Figure 1. A duplicate TCRβ-PCR was performed on 3 different donors. A 1 kb Plus DNA ladder was used. The correct PCR product is ±550 bp long. Note the presence of PCR products other than the desired one, which is a common side effect of SMART cDNA synthesis.
Notes
- The success of this protocol depends on high quality mRNA and a “clean” working environment (DNase/RNase-free and the presence of PCR-grade disposables) throughout the procedure. Contamination must be minimized by cleaning with 10% bleach solution and/or an RNase-inactivator.
- Aside from poor mRNA quality/quantity, many factors can cause the PCR to fail. One of the easier factors to exclude as a cause is the reagents you work with. Make sure all stocks are clean and prepared correctly, using molecular grade water to prepare dilutions. Also, do not expose reagents to room temperature for longer than is necessary.
- Furthermore, use fresh reagents. Especially SuperScript II Reverse Transcriptase is sensitive for losing its activity over time. Ideally, all reagents are less than one year old. While a sample with high input RNA (for example, 1.0 x 106 cells from a T-cell clone) may not suffer from suboptimal reagents, samples with few cells (500-5,000) are very sensitive to the solutions used.
Recipes
- 10x Universal Primer mix
Prepare a mix containing 0.4 μM Universal Primer Long and 2.0 μM Universal primer Short - 50 mM DTT
Dilute the 100 mM DTT stock (from the SuperScript II Reverse Transcriptase kit) 1:1 with molecular grade H2O
Acknowledgments
This protocol was adapted from the previously published studies (Douek et al., 2002; Quigley et al., 2011).
References
- Douek, D. C., Betts, M. R., Brenchley, J. M., Hill, B. J., Ambrozak, D. R., Ngai, K. L., Karandikar, N. J., Casazza, J. P. and Koup, R. A. (2002). A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol 168(6): 3099-3104.
- Koning, D., Costa, A. I., Hoof, I., Miles, J. J., Nanlohy, N. M., Ladell, K., Matthews, K. K., Venturi, V., Schellens, I. M., Borghans, J. A., Kesmir, C., Price, D. A. and van Baarle, D. (2013). CD8+ TCR repertoire formation is guided primarily by the peptide component of the antigenic complex. J Immunol 190(3): 931-939.
- Quigley, M. F., Almeida, J. R., Price, D. A. and Douek, D. C. (2011). Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr Protoc Immunol Chapter 10: Unit10 33.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Nanlohy, N. M., Koning, D., Quakkelaar, E. D. and Baarle, D. V. (2015). TCRβ Clonotype Analysis of EBV and CMV-specific Human CD8+ T Cells. Bio-protocol 5(19): e1606. DOI: 10.21769/BioProtoc.1606.
Category
Immunology > Immune cell function > Lymphocyte
Microbiology > Microbe-host interactions > In vivo model > Mammal
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link