- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of the Cell Proliferation Zone in Leaves by Using EdU
Published: Vol 5, Iss 18, Sep 20, 2015 DOI: 10.21769/BioProtoc.1600 Views: 9636
Reviewed by: Tie LiuFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Efficient Transient Gene Knock-down in Tobacco Plants Using Carbon Nanocarriers
Gozde S. Demirer and Markita P. Landry
Jan 5, 2021 6382 Views

Faster Bacterial Gene Cloning Using the Brick into the Gateway (BiG) Protocol
Flaviani G. Pierdoná [...] Fabio T. S. Nogueira
Dec 20, 2022 2363 Views
Abstract
Visualization of nuclei in S-phase cells in tissues is important for not only cell cycle research but also developmental research because morphogenesis is usually achieved by a combination of cell proliferation and cell expansion. Recently, DNA labeling with 5-ethynyl-2′-deoxyuridine (EdU), which is an analog of thymidine, has been used to visualize nuclei in S-phase cells to assess the activity of cell proliferation during development of plants. EdU is efficiently incorporated into newly synthesized DNA, and detection of EdU is based on the covalent reaction between EdU and Alexa Fluor® dye, which is one of useful fluorescent dyes; this allows us to use mild conditions for the assay without any DNA denaturation. This method could be easily applicable, and, indeed, has been used for various model and non-model plant species. Here, we have described a protocol developed for the detection of nuclei in S-phase cells in leaves.
Keywords: Cell divisionMaterials and Reagents
- Plant tissues
- Click-iT® EdU Alexa Fluor® 488 Imaging Kit (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: c10337 )
- 90 ml acetone (WAKO, catalog number: 019-00353 )
- 1x phosphate-buffered saline (PBS) (e.g. WAKO, catalog number: 314-90185 )
- 0.5% Triton X-100 in 1x PBS
- Plastic tubes (PCR tube or 1.5-ml tube)
- Aluminum foil
- 90% acetone (see Recipes)
- Fixative formalin-acetic acid-alcohol (FAA) (see Recipes)
- 10 mM EdU stock solution (see Recipes)
- 10 µM EdU solution (see Recipes)
- Alexa Fluor® 488 azide solution (see Recipes)
- Click-iT® EdU reaction buffer (see Recipes)
- Click-iT® EdU buffer additive (see Recipes)
- Reaction cocktail (see Recipes)
Equipment
- Microscope (e.g. Nikon, model: ECLIPSE 80i )
- Shaker
- Vacuum pump
- Desiccator
- Razor
- Tweezer
Procedure
- For the experiment, select healthy plants and use seedlings, dissected leaves or leaf primordia, depending on the aim of the experiment. If you need to use older (mature) leaves, cut the leaves using a razor to obtain small-sized leaf discs (Figure 1A).
- Incubate the tissues in PCR tubes filled with 100-200 µl 10 µM EdU solution for 2 to 3 h [Figure 1B; normally 2 h. Kotogány et al. (2010) showed that labeling with 10 µM EdU for 2 h is sufficient for slowly growing cultured cells]. Avoid soaking the tissues or seedlings thoroughly in the solution to ensure that DNA replication and cell division proceed with less stresses, if your plant material is not aquatic plant (Figure 1B).
- After incubation, transfer the tissues gently to 1.5 ml tubes filled with iced 1 ml 90% acetone by tweezers and incubate on ice for 10 min.
- Transfer the tissues gently to other 1.5 ml tubes by tweezers, and wash the tissues 3 times with 1 ml 1x PBS by using a pipette.
- Remove the supernatant and fix the tissues with FAA for 2 to 3 h (the time for fixation depends on the plant materials or tissues). Decompression treatment using a pressure reducing pump for 15 min from the beginning or brief centrifugation are effective for the fixation.
- Transfer the tissues gently to other 1.5 ml tubes by tweezers, and wash the tissues twice with 1 ml 0.5% Triton X-100 in 1x PBS by shaking (100-150 rpm) for 5 min.
- After washing, prepare the reaction cocktail. (Do not prepare the cocktail before washing in order to avoid reducing the activity of the cocktail.)
- Remove the supernatant and add the reaction cocktail to the tubes (generally, 50-100 µl/tube) and mix well by using a pipette.
- Incubate the reaction cocktail for 30 min without any agitation and protect from light by wrapping the tubes with aluminum foil. To enhance the detection sensitivity, repeat step 8 with a fresh 50-100 µl/tube reaction cocktail.
- After incubation, transfer the tissues gently to other 1.5 ml tubes by tweezers and wash the tissues 3 times with 1 ml 1x PBS by shaking (100-150 rpm) for 20 min; protect from light by wrapping the tubes with aluminum foil.
- EdU signals can be observed using a fluorescence microscope (Figure 1C; for the detection of EdU with Alexa Fluor® 488 azide, use 458-488-nm excitation with a green emission filter). EdU signals can be observed a few weeks or even months after the detection reaction if the tissues are stored in 1x PBS in dark, at 4 °C.
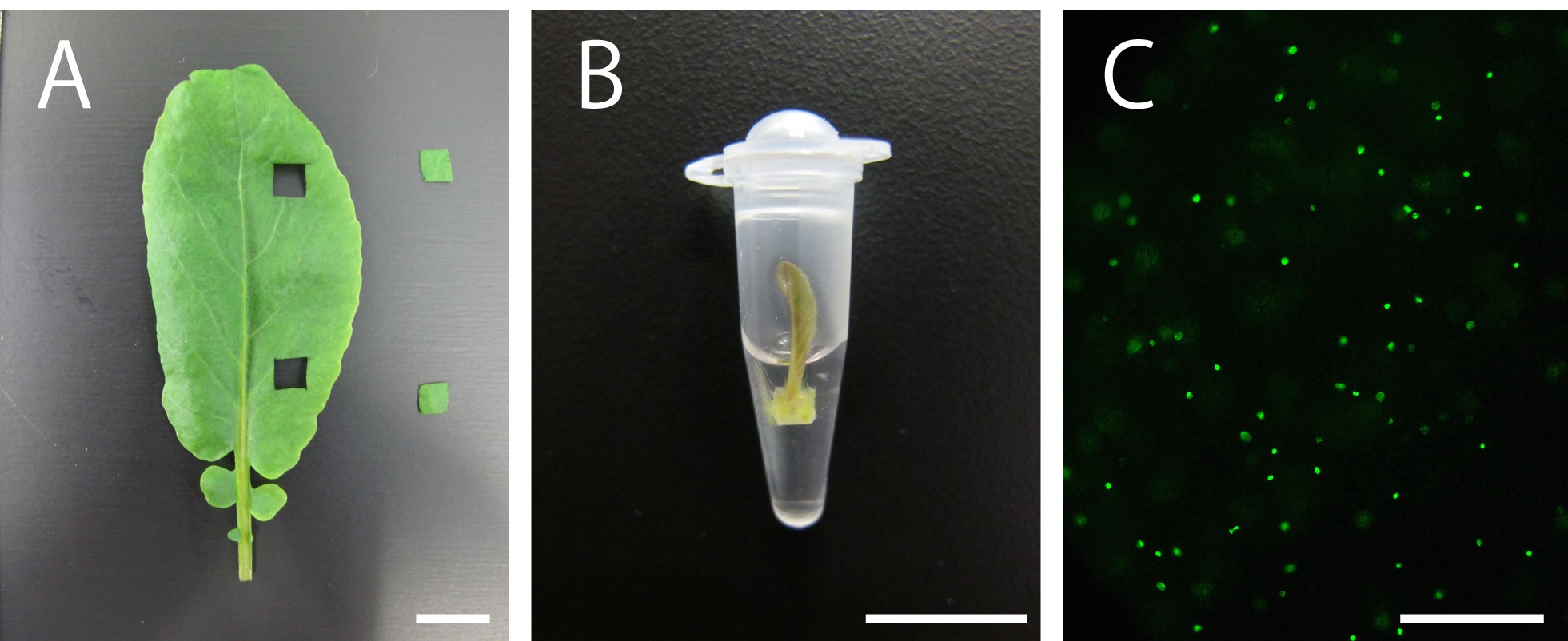
Figure 1. Plant material soaking in 10 µM EdU solution and visualization of nuclei in S-phase cells in the leaves of Rorippa aquatica (Brassicaceae). A. A leaf and leaf discs. B. A shoot apex subtending leaf primordia soaking in 10 µM EdU solution. C. Result of DNA labeling with EdU in leaves. This figure was taken under 20x objective. Bars = 1 cm in (A) and (B) and 100 µm in (C)
Representative data
For representative data, please see the papers of Ichihashi et al. (2014) and Nakayama et al. (2014).
Notes
- Basically, perform all the steps at room temperature.
- In all the steps, work gently to maintain the shape and structure of leaf primordia and leaves.
- Nuclei in S-phase cells can be easily quantified by counting all the signals in a defined area.
- This method is applicable for various plant tissues to detect nuclei in S-phase cells (e.g. shoot apical meristem).
Recipes
- 90% acetone (100 ml)
90 ml acetone
10 ml H2O - FAA (100 ml)
10 ml formaldehyde (37-40%)
5 ml glacial acetic acid
50 ml ethanol
35 ml H2O - 10 mM EdU stock solution
To prepare 10 mM EdU stock solution, add 2 ml of DMSO (Component C) to Component A in the kit and mix well
Stored at -20 °C
When stored as directed, this stock solution will remain stable for up to 1 year - 10 µM EdU solution
To prepare 10 µM EdU solution, dilute 10 mM EdU stock solution to 10 µM by using H2O - Alexa Fluor® 488 azide solution
To prepare a working solution of Alexa Fluor® 488 azide, add 70 µl of DMSO to component B in the kit and mix well
Stored at -20 °C
When stored as directed, this solution will remain stable for up to 1 year - Click-iT® EdU reaction buffer
To prepare Click-iT® EdU reaction buffer, add 3.6 ml of H2O to 400 µl of Click-iT® EdU Reaction Buffer (Component D) in the kit and mix well
Stored at 4 °C
When stored as directed, this 1x buffer will remain stable for up to 6 months - Click-iT® EdU buffer additive
To prepare Click-iT® EdU buffer additive, add 1 ml of H2O to 200 mg of Click-iT® EdU Reaction Buffer additive (Component F) in the kit and mix well
Stored at -20 °C
When stored as directed, this buffer additive will remain stable for up to 1 year - Reaction cocktail
It is important to add the ingredients in the order listed in the table to ensure that the reaction proceeds at an optimal rate
Use the reaction cocktail as soon as possible
Table 1. Reaction cocktail
Acknowledgments
The protocol was modified from Kotogány et al. (2010). We thank Ms. Rumi Amano for preparing the figures. This research was partially supported by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) (KAKENHI Grant Numbers 22870031, 24247007, 24770047 and 25113002) and The Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan to S. K. and BIO-NEXT project from Okazaki Institute for Integrative Bioscience to K. K. and H. T. and by a Research Fellowship from JSPS to H. N..
References
- Ichihashi, Y., Kawade, K. and Tsukaya, H. (2014). Leaf blade and leaf petiole of Arabidopsis thaliana. In: Noguchi, T., Kawano, S., Tsukaya, H., Matsunaga, S., Sakai, A., Karahara, I. and Hayashi, Y. (eds.) Atlas of plant cell structure. Springer, 194-195.
- Kotogany, E., Dudits, D., Horvath, G. V. and Ayaydin, F. (2010). A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6(1): 5.
- Nakayama, H., Nakayama, N., Seiki, S., Kojima, M., Sakakibara, H., Sinha, N. and Kimura, S. (2014). Regulation of the KNOX-GA gene module induces heterophyllic alteration in North American lake cress. Plant Cell 26(12): 4733-4748.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Nakayama, H., Kawade, K., Tsukaya, H. and Kimura, S. (2015). Detection of the Cell Proliferation Zone in Leaves by Using EdU. Bio-protocol 5(18): e1600. DOI: 10.21769/BioProtoc.1600.
Category
Plant Science > Plant developmental biology > Morphogenesis
Plant Science > Plant molecular biology > DNA > DNA modification
Molecular Biology > DNA > DNA labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link













