- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection and Visualization of Specific Gene Transcripts by in situ RT-PCR in Nematode-Infected Arabidopsis Root Tissue
Published: Vol 5, Iss 18, Sep 20, 2015 DOI: 10.21769/BioProtoc.1597 Views: 10351
Reviewed by: Zhaohui LiuDaniel F. CaddellAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Image Analysis Pipeline to Quantify Emerging Cracks in Materials or Adhesion Defects in Living Tissues
Stéphane Verger [...] Olivier Hamant
Oct 5, 2018 6712 Views

Evaluation of Genotoxicity by Micronucleus Assay in vitro and by Allium cepa Test in vivo
Christina N. Banti and Sotiris K. Hadjikakou
Jul 20, 2019 9356 Views

Closed Systems to Study Plant–Filamentous Fungi Associations: Emphasis on Microscopic Analyses
Vasiliki Skiada and Kalliope K. Papadopoulou
Feb 20, 2025 2865 Views
Abstract
This protocol describes an effective method of in situ RT-PCR that was developed to localize specific gene expression directly in thin cross-sections of nematode feeding sites induced by the cyst nematode Heterodera schachtii (H. schachtii) or the root-knot nematode Meloidogyne incognita (M. incognita) in Arabidopsis roots using DIG (Digoxigenin-11dUTP) labeling coupled with AP (alkaline phosphatase) and nitro-blue tetrazolium/5-bromo-4-chloro-3'-indolylphosphate-based detection. This method is applicable to any other Arabidopsis root tissue.
Keywords: In situ RT-PCRMaterials and Reagents
- Arabidopsis roots
Note: Non-infected root fragments or root fragments containing feeding sites of H. schachtii (syncytia) or M. incognita (giant-cells embedded within galls). - Ethanol
- Formaldehyde
- Diethylpyrocarbonate-treated water (DEPC-H2O)
- Low melting agarose
- Petri dishes (
 50 mm)
50 mm) - 12/24-well plates
- Parafilm
- Superglue
- RNase Away (Thermo Fisher Scientific-Molecular BioProducts, catalog number: 7005 )
- DNase I, RNase-free, HC (50 U/µl) (50 U/ul, 1,000 U) ((Thermo Fisher Scientic, Fermentas, catalog number: EN0523 )
- RiboLock RNase Inhibitor (40 U/µl) (RiboLock: 40 U/µl) (Thermo Fisher Scientic, Fermentas, catalog number: EO0381 )
- Ethylenediaminetetraacetic acid (EDTA)
- SuperScriptTM III Reverse Transcriptase Kit (Life Technologies, InvitrogenTM, catalog number: 18080-093 )
- Deoxynucleotides (dNTPs)
- Bovine serum albumin (BSA)
- DIG (Digoxigenin-11dUTP-alkali stable; 25 nmol/25 µl) (Roche Diagnostics, catalog number: 11093088910 )
- BioThermTM Taq DNA Polymerase (GeneCraft, catalog number: GC-002-0100 )
- Anti-Digoxigenin-AP, Fab fragments (150 U/200 µl) (Roche Diagnostics, catalog number: 11093274910 )
- BCIP®/NBT Liquid Substrate System (Sigma-Aldrich, catalog number: B1911-100 ML )
- Fixation solution (see Recipes)
- 10x phosphate buffered saline (PBS) (see Recipes)
- 20x SSC (see Recipes)
- 10x PCR buffer (see Recipes)
- 10x washing buffer (see Recipes)
Equipment
- Vibratome (Leica, model: VT 100 )
- Forceps
- Heating plate
- Laminar flow
- PCR cycler
- Stereo microscope
Procedure
Precautions: RNA is very sensitive and easily degraded by RNases! Therefore always wear gloves, use RNase-free water as well as glass- and plasticware. Recommended is the use of commercially available reagents for eliminating RNase and DNA contamination from the surface of glassware or plasticware e.g. RNase away!
- Fixation
- Cut segments of Arabidopsis roots containing feeding sites of H. schachtii (syncytia) or M. incognita (giant-cells embedded within galls) and corresponding non-infected root fragments.
- Immediately put the root pieces into a microcentrifuge tube containing freshly prepared and pre-chilled fixation solution and vacuum infiltrate it on ice for 15-20 min to ensure the proper penetration of the fixative. Incubate samples 42 h or longer at 4 °C on a shaker with gentle shaking.
- Cut segments of Arabidopsis roots containing feeding sites of H. schachtii (syncytia) or M. incognita (giant-cells embedded within galls) and corresponding non-infected root fragments.
- Embedding
- Subsequently wash fixed samples 3 x 10 min in 63% ethanol in PBS and 1 x 10 min in PBS.
- Embed the tissue in low melting agarose (5% w/v in PBS) in small petri dishes (
 50 mm) placed on a heating plate (35-40 °C) in the laminar flow. Grab the root fragments with the forceps, swab them carefully with a paper towel to remove the excess of PBS and dip them into warm agarose parallel to the bottom of the Petri dish or 12/24-well plate and approx. at the half depth of the agarose. Approx. 10 root segments/dish or 1-2 root segments/12/24-well plate (Figure 1A).
50 mm) placed on a heating plate (35-40 °C) in the laminar flow. Grab the root fragments with the forceps, swab them carefully with a paper towel to remove the excess of PBS and dip them into warm agarose parallel to the bottom of the Petri dish or 12/24-well plate and approx. at the half depth of the agarose. Approx. 10 root segments/dish or 1-2 root segments/12/24-well plate (Figure 1A). - Cool down the agarose in the laminar flow.
- Petri dishes sealed with Parafilm can be stored in the fridge (4 °C) for several weeks.
- Subsequently wash fixed samples 3 x 10 min in 63% ethanol in PBS and 1 x 10 min in PBS.
- Preparation of cross sections with the vibratome and DNase treatment
- Cut out small agarose blocks (approx. 8 mm high x 5 mm wide) containing the syncytia or non-infected root segments with a scalpel. Trim the agarose block under the microscope (Figure 1B) and glue it on the round vibratome plate with a drop of common superglue. To produce cross sections the root segments within the agarose block have to be placed perpendicular to the bottom of the vibratome plate (Figure 1B-C).
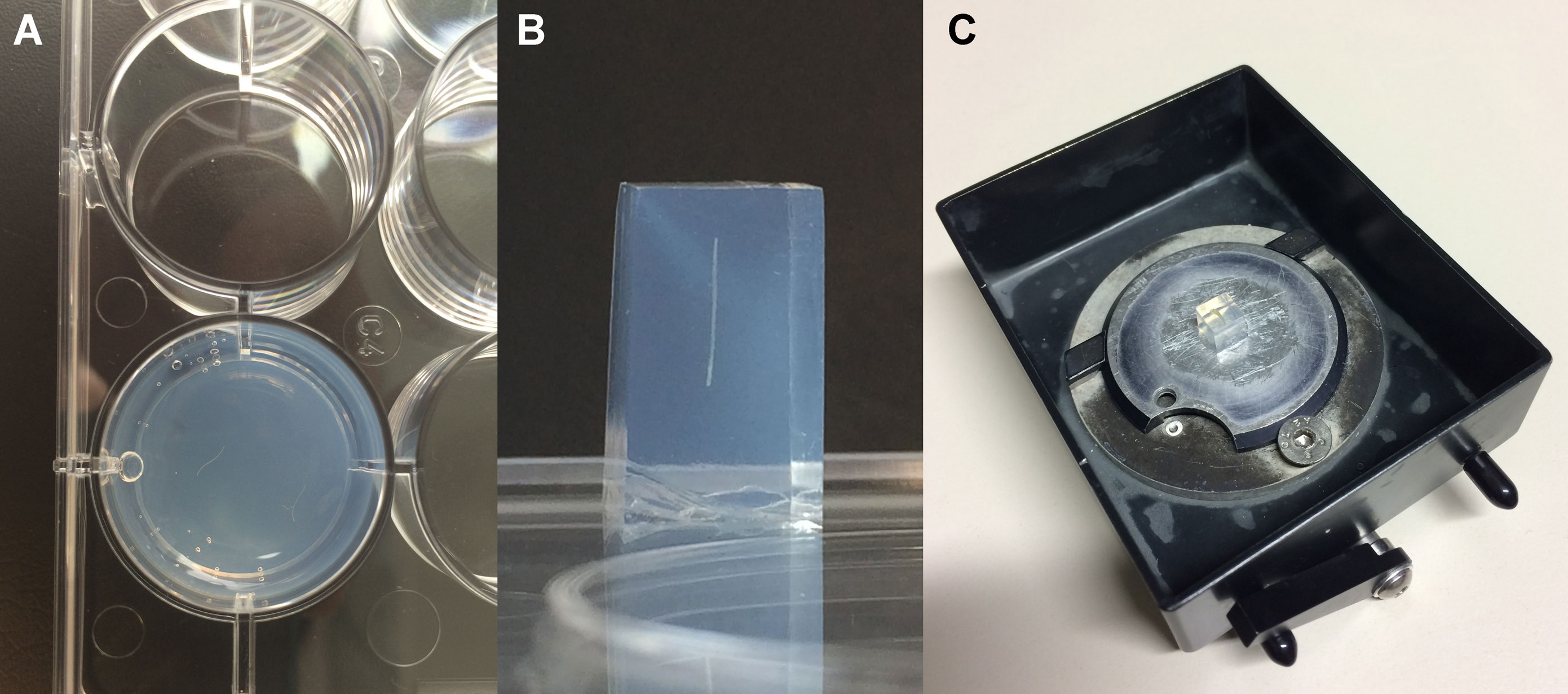
Figure 1. Placement of the root fragments within a well of the 12-well plate (A; view from above), trimmed agarose block with a root fragment in the perpendicular position (B; view from the side) and agarose block glued onto the vibratome plate (C) - Cut 20-25 µm thick tissue sections in the vibratome tray filled with DEPC-H2O.
Using forceps (in case the tissue section remains in the agarose section) or a pipette (in case the tissue section disconnects from the agarose section) collect the sections in a watchglass or similar container filled with DEPC-H2O supplemented with RNase inhibitor. - With a pipette remove the excess of DEPC-H2O (as much as possible). To prevent non-specific DNA signals digest the DNA within the tissue by adding 2 µl RNase-free DNAse and 1 µl RNase Inhibitor to the remaining DEPC-H2O covering the cross-sections. Subsequently incubate the samples overnight at 37 °C in a watchglass in a Petri dish sealed with parafilm.
- Cut out small agarose blocks (approx. 8 mm high x 5 mm wide) containing the syncytia or non-infected root segments with a scalpel. Trim the agarose block under the microscope (Figure 1B) and glue it on the round vibratome plate with a drop of common superglue. To produce cross sections the root segments within the agarose block have to be placed perpendicular to the bottom of the vibratome plate (Figure 1B-C).
- Washing
- Add several drops of 0.5 M EDTA and incubate the cross-sections at 65 °C for 10 min to inactivate the DNase.
Subsequently wash sections as follows:
2 times with 2x SSC buffer
1 time with 1x SSC buffer
1 time with 0.5x SSC buffer
2 times with ddH2O
Each step should be performed with 1 ml of buffer or ddH2O in a sealed Petri dish at 37 °C for 10 min.
- Add several drops of 0.5 M EDTA and incubate the cross-sections at 65 °C for 10 min to inactivate the DNase.
- Reverse transcription
- Transfer several sections into a PCR tube using a pipette (in ca. 11.5 µl ddH2O).
- Add 0.5 µl 3’ primer and 1.0 µl dNTPs and start the first step of the RT reaction: 65 °C for 5 min.
- Cool down the samples, incubate them at least 1 min on ice and add 4 µl of the 5x First-Strand Buffer, 1 µl 0.1 M DTT, 1 µl RNase Inhibitor and 1 µl SuperScriptTM Reverse Transcriptase. Total volume 20 µl.
- Run the following program in a PCR cycler:
25 °C - 5 min
55 °C - 1 h
75 °C - 5 min (inactivation of DNase)
- Transfer several sections into a PCR tube using a pipette (in ca. 11.5 µl ddH2O).
- PCR
- Prepare a dNTPs mix containing 10 mM dGTP, dATP and dCTP. Add to the RT-mix as follows:
5.0 µl - 10x PCR buffer
1.0 µl - 3’ primer (final concentration 0.5 µM)
1.0 µl - 5’ primer (final concentration 0.5 µM)
1.0 µl - dNTPs mix
2.36 µl - 2 mM dTTP
0.5 µl - DIG
0.25 µl - BioThermTM Taq DNA Polymerase
18.89 µl - ddH2O
Total volume 50 µl (30 µl of PCR-mix and 20 µl of RT-mix).
In case of the negative control the polymerase will be omitted.
- Prepare a dNTPs mix containing 10 mM dGTP, dATP and dCTP. Add to the RT-mix as follows:
- Washing and detection
The following steps are carried out under the stereo microscope at room temperature (from step G5 in dark room with a weak source of light e.g. small table lamp). Short centrifugation could be necessary between each washing step to collect the sections at the bottom of the PCR tube. This procedure might prevent the accidental removal of tissue sections during several washing rounds. You might also check the discarded liquids for unintended removed sections after each washing step.- Wash the samples twice in 1x PBS buffer for 5 min.
- To block unspecific binding sites incubate the sections for 30 min in freshly prepared blocking solution containing 0.1% w/v BSA in 1x PBS.
- Remove the blocking solution and add 50 µl Anti-DIG-AP, Fab fragments diluted 1:500 in blocking solution. Incubate 1 h at RT.
- Remove the blocking solution containing Anti-DIG-AP, Fab fragments and wash the sections 2 x 15 min with 10x washing buffer.
- Using a pipette place a drop of the washing buffer containing section(s) on a microscopic slide and replace the washing buffer with 50 µl of the NBT/BCIP solution (1:50 in washing buffer). Keep the slide in darkness (use e.g. lid from chemical bottle as a cover) since the NBT/BCIP is light sensitive!
- From time to time observe the progress of the staining reaction under the microscope, it appears usually within first 2-3 min (keep the light exposure as short as possible).
- If the violet staining is clearly visible stop the reaction with ddH2O and photograph your sections. Figure 2 shows an example of an in situ RT-PCR analysis made for two β-1, 4- endoglucanases, AtCel1 and KOR3, which are specifically up-regulated in syncytia induced by H. schachtii in Arabidopsis roots.
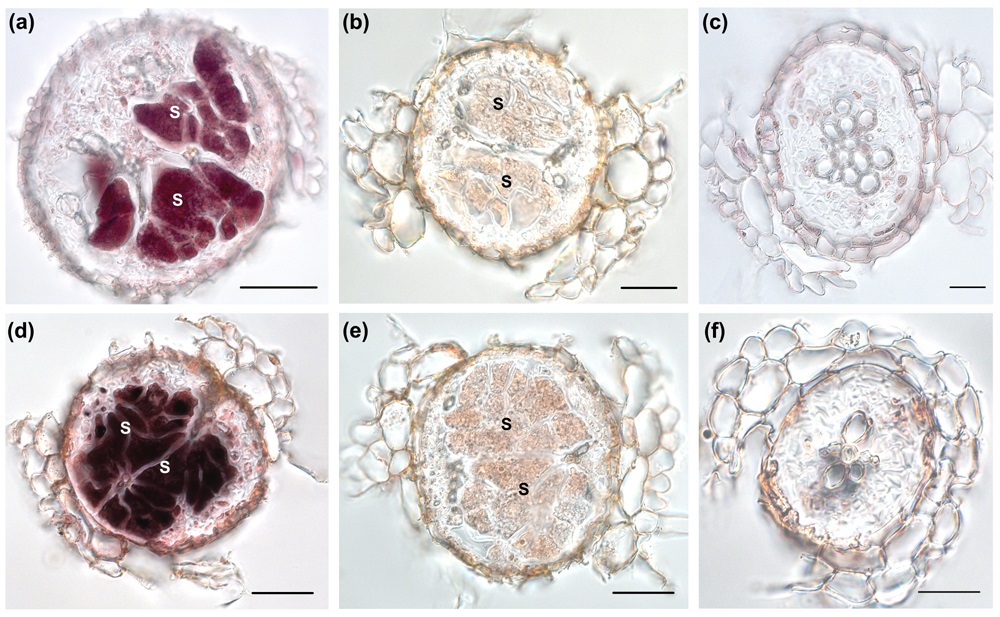
Figure 2. In situ RT-PCR analysis of AtCel2 and KOR3 on sections of syncytia induced by Heterodera schachtii at 10 days after inoculation (dai). (a) AtCel2 transcripts accumulate within syncytium. (b) Control reaction for (a) performed without polymerase. Staining is not detected. (c) Control reaction for (a) on a root section above the syncytium. Transcripts of AtCel2 were not detected. (d) Strong staining associated with transcripts of KOR3 is visible in the syncytium. (e) Control reaction to (d) performed without polymerase; no staining is visible. (f) Control reaction for (d) on a root section above the syncytium. No transcripts of KOR3 are visible. (a), (b), (d) and (e) scale bar = 50 µm; (c) and (f) scale bar = 20 µm. From Wieczorek et al. (2008), with permission.
- Wash the samples twice in 1x PBS buffer for 5 min.
Notes
- RNA is easily degraded by RNAses and several different feeding sites/roots should be used to enhance the success rate of this method. However, if the feeding site/root tissue that is used contains intact RNA the reproducibility of this protocol is high and the variability low.
- Start the detection with the positive reaction and subsequently use exactly the same exposure time for the controls.
- The coloration of gene transcript-specific staining may vary from light violet to dark violet (almost black). The unspecific staining in the control sections (without Taq Polymerase) may vary from light brown, reddish to very faint/almost transparent.
Recipes
- Fixation solution
63% ethanol
5% formaldehyde in PBS (containing DEPC-ddH2O, pH 7.2) - 10x phosphate buffered saline (PBS)
10 mM Na2HPO4
130 mM NaCl in DEPC-ddH2O
pH 7.5 - 20x SSC
3 M NaCl
300 mM C6H5O7.2H2O.3Na (sodium citrate dihydrate) in DEPC-ddH2O - 10x PCR buffer
160 mM (NH4)2SO4
670 mM Tris-HCl (pH 8.8) (at 25 °C)
15 mM MgCl2
0.1% Tween 20
5.10x washing buffer
0.1 M Tris-HCl
0.15 M NaCl
pH 9.5
Acknowledgments
This protocol is a combination of methods previously published by Koltai and Bird (2000) and Urbanczyk-Wlochniak et al. (2002) and has been additionally modified. This work was supported by grant QLK-CT-1999-01501 (‘NONEMA’) from the European Union within the 5th Framework and FWF grants P16296-B06, P16897-B06 and P21067-B12.
References
- Hofmann, J., Wieczorek, K., Blochl, A. and Grundler, F. M. (2007). Sucrose supply to nematode-induced syncytia depends on the apoplasmic and symplasmic pathways. J Exp Bot 58(7): 1591-1601.
- Koltai, H. and Bird, D. M. (2000). High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol 123(4): 1203-1212.
- Siddique, S., Endres, S., Atkins, J. M., Szakasits, D., Wieczorek, K., Hofmann, J., Blaukopf, C., Urwin, P. E., Tenhaken, R., Grundler, F. M., Kreil, D. P. and Bohlmann, H. (2009). Myo-inositol oxygenase genes are involved in the development of syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytol 184(2): 457-472.
- Urbanczyk-Wochniak, E., Filipecki, M. and Przybecki, Z. (2002). A useful protocol for in situ RT-PCR on plant tissues. Cell Mol Biol Lett 7(1): 7-18.
- Wieczorek, K., Golecki, B., Gerdes, L., Heinen, P., Szakasits, D., Durachko, D. M., Cosgrove, D. J., Kreil, D. P., Puzio, P. S., Bohlmann, H. and Grundler, F. M. (2006). Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J 48(1): 98-112.
- Wieczorek, K., Hofmann, J., Blochl, A., Szakasits, D., Bohlmann, H. and Grundler, F. M. (2008). Arabidopsis endo-1,4-beta-glucanases are involved in the formation of root syncytia induced by Heterodera schachtii. Plant J 53(2): 336-351.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wieczorek, K. (2015). Detection and Visualization of Specific Gene Transcripts by in situ RT-PCR in Nematode-Infected Arabidopsis Root Tissue . Bio-protocol 5(18): e1597. DOI: 10.21769/BioProtoc.1597.
Category
Microbiology > Microbe-host interactions > Nematode
Plant Science > Plant cell biology > Tissue analysis
Cell Biology > Cell staining > Nucleic acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










