- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Respiratory Activity in Biofilms
Published: Vol 5, Iss 18, Sep 20, 2015 DOI: 10.21769/BioProtoc.1591 Views: 10797
Reviewed by: Maria SinetovaAlexander B. WestbyeEsteban Paredes-Osses

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Inexpensive Imaging Platform to Record and Quantitate Bacterial Swarming
Weijie Chen [...] Jay X. Tang
Sep 20, 2021 3643 Views

Purification of the Bacterial Amyloid “Curli” from Salmonella enterica Serovar Typhimurium and Detection of Curli from Infected Host Tissues
Murugesan Sivaranjani [...] Aaron P. White
May 20, 2022 3236 Views

A Guideline for Assessment and Characterization of Bacterial Biofilm Formation in the Presence of Inhibitory Compounds
Bassam A. Elgamoudi and Victoria Korolik
Nov 5, 2023 3130 Views
Abstract
Bacteria live mostly as biofilms, not as planktonic cell populations. Bacterial cells living as biofilms are known to be in different physiological status. Persister cells are one of such physiological conditions and they are recognized as to be a stochastically produced sub-population of non-growing bacterial cells. The following protocol describes a method to determine the respiratory activity of cells within biofilms.
Materials and Reagents
- Bacterial culture [Pseudomonas aeruginosa (P. aeruginosa) PA14]. This method has also been performed for Escherichia coli (BW25113)
- LB broth Lennox (BD, catalog number: 240230 )
- CorningTM cellgroTM Ciprofloxacin Hydrochloride (Thermo Fisher Scientific, catalog number: 61-277-RF )
- BacLightTM RedoxSensorTM CTC Vitality Kit (Thermo Fisher Scientific, catalog number: B34956 )
- SYTO® 40 Blue Fluorescent Nucleic Acid Stain-5 mM Solution in DMSO (Thermo Fisher Scientific, catalog number: S11351 )
- Sodium chloride (VWR International, BDH®, catalog number: 0241-VBD )
- Aluminum Foil
- Spooled Masterflex peroxide-cured silicone tubing, L/S 14, 250 ft. (Cole-Parmer, catalog number: UX-96407-14 )
- Masterflex Norprene tubing (A60 G), L/S 13, 50 ft. (Cole-Parmer, catalog number: UX-06404-13 )
- PVDF barbed Y connector, 3/8" ID, 1/4", 2-3/8", 1-3/4"; Pack Of 10 (Cole-Parmer, catalog number: WU-30703-93 )
- Barbed fittings, Straight Connector, Clear PP,1/16" ID, 1/32", 25/32", 1/4" (Cole-Parmer, catalog number: WU-30506-00 )
- Barbed fittings, Reducing Connector, Clear PP,1/8" x 1/16" ID, 3/32", 13/16", 1/8" (Cole-Parmer, catalog number: WU-30506-06 )
- Barbed fittings, T connector, Kynar, 1/4" ID, 1/8", 1-15/16", 1-5/16"; 10/pack (Cole-Parmer, catalog number: WU-30703-75 )
- Micropipettes (P1000, P200, P20)
- Syringe 10 ml
- Syringe needle
- Glass microscope slides (25 x 75 x 1 mm)
- Glass microscope coverslips (60 x 24 mm; no 2)
- Saline (see Recipes)
Equipment
- Peristaltic pump (Cole-Parmer, catalog number: EW-07553-80) with an 8 channel, 6 rollers, 3-stop Ismatec minicartridge pump head (Cole-Parmer, catalog number: EW-78002-50 )
- Inoculation ports (VWR® Sleeve Stoppers) (VWR International, catalog number: 89097-534 )
- GeneMate Incubated Shakers (BioExpress, catalog number: H-2000-M )
- Nalgene® Carboys with Handles, Polypropylene, Thermo Scientific (VWR International, catalog number: 16101-084 )
- Nalgene® Top WorksTM Aseptic Closure System, Silicone, for Bottles and Carboys, Thermo Scientific (VWR International, catalog number: 2135-8303 )
- Acro® 50 Vent Filters, Pall Laboratory (VWR International, catalog number: 28143-616 )
- Anodized Transmission flow cell reactors (Biosurface technologies, catalog number: FC 81-Al )
- Confocal Scanning Microscope and Analysis software (e. g. Leica Confocal SP5 Imaging system and Software)
Software
- COMSTAT software (http://www.comstat.dk)
- Intensity Luminance V1 software (http://bingweb.binghamton.edu/~scraver/IL.html)
- ImageJ software (http://imagej.nih.gov/ij/)
- Leica LAS AF software (Leica)
Procedure
The procedure below, describes how to assess the respiratory activity of persister cells present within biofilms. However, the method can be adapted to overall biofilm populations. In this method, differential staining is used where respiratory activity (metabolic activity is assessed) using the monotetrazolium redox dye 5-cyano-2, 3-ditolyl tetrazolium chloride (CTC) that produces a fluorescent formazan (CTF, indicated by cells stained in red) when reduced. All cells are stained with the nucleic acid stain SYTO 40.
- Biofilm development in flow cell reactors.
- Prepare a streak plate on LB agar of a frozen stock of the bacteria of interest (in this case Pseudomonas aeruginosa PA14) and incubate under static conditions at 37 °C, for 24 h.
- Remove 2 colonies from the streak plate and inoculate a broth culture of 5 ml LB broth.
- Incubate at 37 °C with agitation (220 rpm) for a period of 12-24 h.
- Dilute the overnight culture to 1% in 50 ml of fresh LB medium in an Erlenmeyer flask.
- Incubate at 37 °C with agitation (220 rpm) for a period of 24 h.
- Measure the OD600 of the overnight culture and standardize the culture to have an OD600 of 0.8 in 10 ml of LB.
- Aspirate the culture with a 10 ml syringe with a needle and then cap the needle.
- Inoculate each flow cell reactor.
- In this example, biofilms were cultured in flow cell reactors (Figure 1), as described previously (Sauer et al., 2002; Davies and Marques, 2009; Marques et al., 2014). Each flow cell reactor was inoculated with 3 ml of a standardized overnight culture (OD600 of 0.8) and incubated, under static conditions, for a period of 1 h, to facilitate cell attachment.
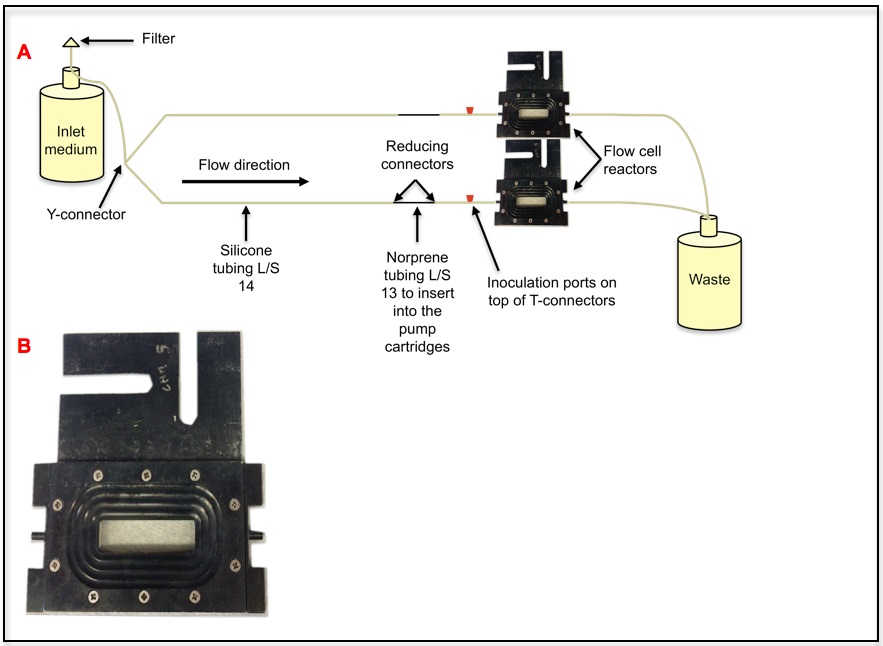
Figure 1. Flow cell reactor system. This is a once through continuous culture system. Once the bacterial culture is inoculated into the inoculation port, biofilms develop on the coverslip of the flow cell reactor. Only the bottom reactor is labeled, the top reactor is set in an identical manner. One of the flow cell reactor sets is labeled. A. Schematic version of a flow cell system where 2 flow cells are run together. B. Flow cell reactor.
- In this example, biofilms were cultured in flow cell reactors (Figure 1), as described previously (Sauer et al., 2002; Davies and Marques, 2009; Marques et al., 2014). Each flow cell reactor was inoculated with 3 ml of a standardized overnight culture (OD600 of 0.8) and incubated, under static conditions, for a period of 1 h, to facilitate cell attachment.
- Following 1 h incubation at room temperature - to allow the bacteria to attach to the coverslip (this time can vary, depending on the bacterial strain) - initiate the flow.
- Allow biofilms to develop for a period of 6 days at room temperature (time and temperature may vary, depending on bacterial species used).
- At day 6 expose the P. aeruginosa PA14 biofilms to ciprofloxacin (150 x MIC) or saline (control) for a period of 18 h.
Note: When assessing overall mature biofilm populations, not persister cell populations, at day 6 perform step 12. Respiratory activity can also be assessed at different days, during biofilm development.
- Expose the remaining biofilms to saline containing SYTO 40 (5 μM) and CTC (5 mM), for a period of 60 min. CTC has previously been used to determine the respiratory activity of bacteria within biofilms (Schaule et al., 1993).
- When attempting to determine the effects of certain compounds on persister cell activity, perform this step for 30 min only. The treatment to be performed should be initiated following this step.
- Take several z-stack images of biofilm cell clusters that have developed on the flow cell coverslip by confocal scanning laser microscopy (CSLM) using the following excitation/emission parameters:
- SYTO 40: ex 405, em 450-455
- CTC: ex 476, em 650-680.
- If performing step 12a, then following 30 min, expose the biofilm population to saline or the testing agent, together with SYTO 40 (5 μM) and CTC (5 mM) for further 60 min.
- SYTO 40: ex 405, em 450-455
- Quantify relative fluorescence with the program Intensity Luminance V1 software. This program determines the relative fluorescence present on the images by quantifying the green, blue and red fluorescence. We have provided a link for the software, which contains the instructions of how to use it; we developed the program and made it available to other researchers as freeware.
- SYTO 40 (blue) will stain all cells while CTC stain (red) will stain only respiratory active cells. Thus, 100% of the cells will stain blue, making it possible to determine which percentage of the population is undergoing respiratory activity (red cells) during a 60 min period. If steps 12a and 13c were performed then, the results obtained for the initial 30 min should be used as baseline. In the case of persister cells, the difference between persister cells and overall population would be assessed.
- Perform quantitative analysis of biofilms using the COMSTAT software (Heydorn et al., 2000). A newer version of the COMSTAT software is now available, and has a plugin to enable its use with ImageJ.
- This program is widely used by biofilm researchers to determine and quantify: total biofilm biomass, biofilm cluster thickness, portion of slice occupied by the bacteria, surface area of biomass and surface to biovolume ratio. Blue channel (total) and red channel (respiratory active) images can be analyzed, and differences in respiratory activity between controls and treatments can be performed. In the case of persister cells, the difference between persister cells and overall population would be assessed.
- To assess the respiratory activity, compare between total cells (SYTO 40) and CTC stained cells (respiratory active cells).
- Repeat the experiment 3-4 times to ensure reproducible data and to allow for calculation of whether the results obtained are statistical significant when comparing controls to treated samples using ANOVA, followed by the Tukey’s comparison test.
- Prepare a streak plate on LB agar of a frozen stock of the bacteria of interest (in this case Pseudomonas aeruginosa PA14) and incubate under static conditions at 37 °C, for 24 h.
Representative data
Our laboratory has made use of this type of assay to assess whether the fatty acid signaling molecule cis-2-decenoic acid (cis-DA) would increase the respiratory activity of P. aeruginosa persister cells present in biofilms (Marques et al., 2014). Biofilms were cultured as described in the procedure above, followed by exposure to ciprofloxacin (150 mg/L) for a period of 24 h. Subsequently, the remaining biofilm population, consisting of solely persister cells, was exposed to saline containing SYTO 40 and CTC for a period of 30 min (to obtain a baseline for the stain measurements) followed by a 1 h exposure to either saline or cis-DA (100 nM) in saline. Biofilms were visualized by confocal scanning laser microscopy (Leica Confocal TCS SP5 Imaging System with a DMI 6000 inverted microscope) using the parameters described in the procedure above. Images were acquired with the Leica LAS AF software. In the presence of cis-DA, a significant increase of CTC stain (red panel) was observed compared to exposure to saline alone (Figure 2A) consisting of a 10% increase (P<0.0001). We also observed that persister cells display a respiratory activity, which is indicative of an active metabolic status, albeit being 2.5% (Figure 2B), a low level not visually observed (Figure 2A).
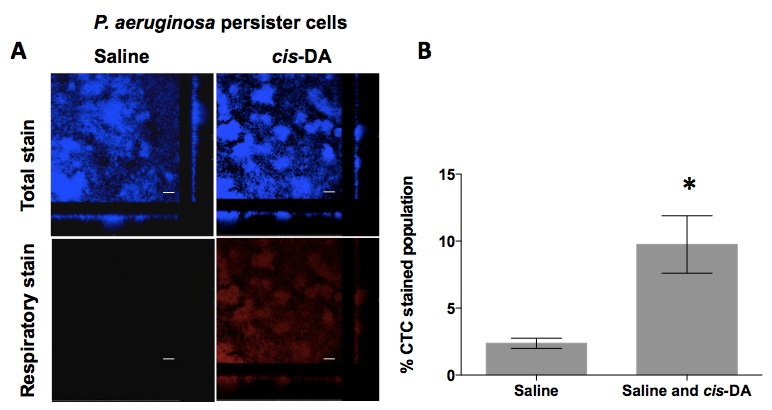
Figure 2. Respiratory activity of P. aeruginosa PA14 persister cells upon exposure to cis-DA. A. Respiratory activity was evaluated by the cell’s ability to metabolize CTC into a fluorescent formazan, following 1 h incubation with saline or cis-DA in saline. SYTO 40 was used to stain overall population; bar equals 25 μm. B. The percentage of CTC stained population of persister cells compared to SYTO 40 stained population was calculated. Error bars indicate standard deviation. *, P<0.001 - Significantly different from persister cells treated with saline alone, as indicated by one way ANOVA. [Copyright© 2014, American Society for Microbiology (Marques et al., 2014)]
Notes
Images of background measurements, using confocal microscopy, should be acquired 25 min following the initial exposure of biofilms to CTC and SYTO 40. Second exposure of to CTC and SYTO 40 should only be initiated once all the background images were acquired.
Recipes
- Saline
8.5 g sodium chloride
1 L of distilled water (DI)
Acknowledgments
The respiratory activity assay using CTC and SYTO 40 is a modification of previously published protocols (Schaule et al., 1993). COMSTAT analysis is based on previously published software (Heydorn et al., 2000). This work was supported by SUNY structural funds.
References
- Davies, D. G. and Marques, C. N. (2009). A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191(5): 1393-1403.
- Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersboll, B. K. and Molin, S. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146: 2395-2407.
- Marques, C. N., Morozov, A., Planzos, P. and Zelaya, H. M. (2014). The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl Environ Microbiol 80(22): 6976-6991.
- Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W. and Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184(4): 1140-1154.
- Schaule, G., Flemming, H. C. and Ridgway, H. F. (1993). Use of 5-cyano-2,3-ditolyl tetrazolium chloride for quantifying planktonic and sessile respiring bacteria in drinking water. Appl Environ Microbiol 59(11): 3850-3857.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Marques, C. N. H. and Craver, S. A. (2015). Quantification of Respiratory Activity in Biofilms. Bio-protocol 5(18): e1591. DOI: 10.21769/BioProtoc.1591.
Category
Microbiology > Microbial biofilm > Biofilm culture
Microbiology > Microbial cell biology > Cell staining
Microbiology > Microbial physiology > Respiration
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










