- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Non-Radioactive Method for Measuring PP2A Activity in Plants
Published: Vol 5, Iss 17, Sep 5, 2015 DOI: 10.21769/BioProtoc.1577 Views: 9060
Reviewed by: Tie LiuHarrie van ErpAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1474 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3209 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views
Abstract
Protein phosphatase 2A (PP2A) is a group of important cellular regulators in eukaryotes that dephosphorylate more than 30% of cellular proteins whose activities are turned on or off by phosphorylation. In plants, PP2A was found to regulate critical components involved in plant growth and development, and in response to biotic and abiotic stresses. Therefore, determining the PP2A activities at different developmental stages, in different tissues, or in various mutants is critical in order to understand the functions of PP2A in plants. Traditional PP2A enzyme assay uses radioactive isotope and often take days to finish. This PP2A enzyme assay described here is a method to determine PP2A activity without using radioactive materials in less than 6 h.
Keywords: PP2A activityMaterials and Reagents
- Serine/Threonine Phosphatase Assay Kit (Promega Corporation, catalog number: V2460 )
- Protein Assay Dye Reagent (Bio-Rad Laboratories, AbD Serotec®, catalog number: 500-0006 )
- Protein Phosphatase (PPase) Inhibitor 2 (I-2) (New England BioLabs, catalog number: P0755S )
- Liquid nitrogen (N2)
- Tris (pH 7.0) (Thermo Fisher Scientific, catalog number: BP152-5 )
- EDTA (Thermo Fisher Scientific, catalog number: S312-12 )
- DTT (Sigma-Aldrich, catalog number: D0632 )
- Brij 35 (Thermo Fisher Scientific, catalog: BP345-500 )
- 1x PP2A assay buffer (see Recipes)
Equipment
- Tabletop centrifuge for 96-well plate (Eppendorf, model: 5810R )
- Centrifuge at 4 °C or in cold-room (Eppendorf, model: 5415D )
- Microplate reader (xMark™ Microplate Absorbance Spectrophotometer) (Bio-Rad Laboratories, catalog number: 1681150 )
- Pestle and mortar
- 37 °C waterbath
Procedure
- Preparation of the desalting column (Note 1)
- Pre-wet the spin column provided in the kit with sterile water.
- Transfer 10 ml of G25 solution from Serine/Threonine Phosphatase Assay kit to each spin column and let G25 settle by gravity for 5 min.
- Wash the column 3 times with 10 ml sterile water, let G25 settle by gravity. Don’t let G25 dry-out.
- Wash the column 3 times with 10 ml of 1x PP2A assay buffer, let G25 settle by gravity, seal the column with a cap and proceed to step B1.
- Put the column into an empty 50 ml centrifuge tube and spin at 600 x g for 5 min at 4 °C.
- Pre-wet the spin column provided in the kit with sterile water.
- Isolate and purification of cellular extracts
- Freeze plant materials in liquid N2, if not used immediately, store frozen samples in -80 °C freezer.
- Grind frozen plant samples in liquid N2 to fine powder in a pre-cooled mortar and transfer powder to 1.5 ml Eppendorf tubes.
- Add 100 µl 1x PP2A assay buffer to each sample (100 mg powder), vortex vigorously to thaw the sample and incubate on ice for 30 min.
- Centrifuge at 13,000 rpm for 30 min at 4 °C.
- Transfer the supernatant to a fresh tube and incubate the crude extracts on ice briefly.
- Proceed to step A5 to prepare the desalting column.
- Transfer the supernatant in step B5 to a prepared desalting column in step A5.
- Centrifuge the samples at 600 x g for 5 min at 4 °C.
- Collect the flow through fluid into fresh tubes and keep them on ice.
- Measure the protein concentration with the Bio-Rad Protein Assay Dye Reagent by mixing 200 µl dye, 800 µl water and 2 µl extracts followed by incubation at room temperature for 5 min before OD595 is recorded, and then normalize the protein concentration to 1 µg/µl with 1x PP2A assay buffer (Protein concentration=6.8*OD595 µg/µl).
- Freeze plant materials in liquid N2, if not used immediately, store frozen samples in -80 °C freezer.
- Constructing a standard curve for determining the amount of free phosphate
- Prepare tubes containing 0, 100, 200, 500, 1,000, 2,000 pmol of phosphates from the 10 mM standard provided in the Serine/Threonine Phosphatase Assay Kit. To ensure the data are comparable, the liquid in all tubes should have a final volume of 50 µl.
- Prepare the Molybdate dye/Additive mix (10 µl Additive into 1 ml Molybdate dye) provided by the kit. Use 50 µl Dye/Additive for each tube and mix by vortexing.
- Centrifuge at 13,000 rpm at room temperature for 30 sec.
- Carefully transfer the supernatants to the 96-well plate provided by the kit while avoiding air-bubbles.
- Centrifuge the 96 well plate for 2 min followed by incubation at room temperature for 5 min.
- Read the optical density with a plate reader using a 600 nm filter.
- Prepare the standard curve based on the OD600 readings and the pmol phosphates used in the assay (Note 5).
- Prepare tubes containing 0, 100, 200, 500, 1,000, 2,000 pmol of phosphates from the 10 mM standard provided in the Serine/Threonine Phosphatase Assay Kit. To ensure the data are comparable, the liquid in all tubes should have a final volume of 50 µl.
- The PP2A activity assay
- Prepare the Molybdate dye/Additive just before the assay starts. Use 50 µl Dye/Additive for each PP2A assay. Calculate the amount needed for the experiment and only prepare sufficient fresh solution for each assay (Note 6).
- Mix the PP2A assay components on ice.
Components
Amount
Purified extracts
5 µl
Peptide substrates
5 µl
10x PP2A assay buffer
5 µl
PPase inhibitor 2
1 µl
Phosphate free water
34 µl
- Incubate the reaction mixture at 37 °C for 5 min.
- Terminate the reaction by adding 50 µl Dye/Additive.
- Mix well and centrifuge at 13,000 rpm at room temperature for 1 min.
- Transfer the mixture to a 96-well plate and avoid air-bubbles (Note 7).
- Allow the plate to incubate at room temperature for 15 min for color development (Note 8).
- Read the optical density with a plate reader using a 600 nm filter.
- Prepare the Molybdate dye/Additive just before the assay starts. Use 50 µl Dye/Additive for each PP2A assay. Calculate the amount needed for the experiment and only prepare sufficient fresh solution for each assay (Note 6).
- Calculation of PP2A activity
- Based on the standard curve, determine the amount of phosphate released from the reaction.
- Use the following equation to determine the PP2A activity.
PP2A activity = amount of phosphate released (pmol)/5 (min)/5 (µg).
- The PP2A activity unit is defined as pmol phosphate released from substrates per min per µg total protein. The PP2A activity should be around 55-60 units for cellular extracts from wild-type Columbia Arabidopsis plants.
- Technical replicates and biological replicates should be performed to obtain the variance between experiments.
- Based on the standard curve, determine the amount of phosphate released from the reaction.
Representative data
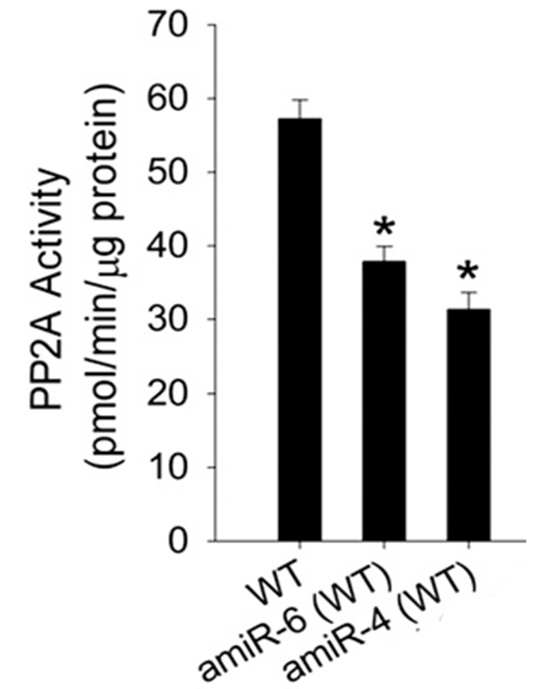
Figure 1. The PP2A activity of WT and two plants (amiR-6 & amiR-4) containing artificial microRNAs designed to down regulate AtPTPA. A Typical assay for WT Arabidopsis has PP2A activity of 55~60 units. amiR plants have reduced PP2A activity due to the degradation of mRNA of AtPTPA, which is essential for PP2A holo-enzyme assembly (see Chen et al., 2014 for detail).
Notes
- It is important that the protein samples are desalted in order to remove the free phosphate that can interfere with the measurements of the phosphate released by PP2A.
- Samples need to be completely thawed and mixed well before centrifugation at 4 °C.
- Avoid transferring debris, as it will clog the desalting column.
- Use x g force in centrifugation; however, if using microfuge, rpm is used in desalting the crude extracts, higher speed gives varied desalting efficiency and sample yield.
- Construct a new standard curve every month after the kit is opened. Please refer the short manual included in this kit for more information about the standard curve. Link to the manual.
- All the assay component should be stored at 4 °C. Never freeze the peptide substrates and the kit should be stable for 3 months after it is opened.
- Spin the 96-well plate again to remove the air bubble if bubble occurs.
- If higher amount of protein (greater than 5 µg) is used for the assay, allow more time for color development. Be sure that the amount of PP2A in the extract does not deplete the phosphate group from the peptide substrates. Perform a titration experiment to determine a linear range of reactions, so the actual assay can be performed within linear range of reactions.
Recipes
- 1x PP2A assay buffer
50 mM Tris (pH 7.0)
0.1 mM EDTA
5 mM DTT and 0.01% Brij 35
Acknowledgments
This work was supported by the Department of Biological Sciences and the Graduate School of Texas Tech University, by the National Natural Science Foundation of China (grant no. 31170793 to Guoxin Shen), and by the National Natural Science Foundation of Zhejiang Province (grant no. Z12c130011 to Guoxin Shen). The protocol is adapted from Chen et al. (2014).
References
- Chen, J., Hu, R., Zhu, Y., Shen, G. and Zhang, H. (2014). Arabidopsis PHOSPHOTYROSYL PHOSPHATASE ACTIVATOR is essential for PROTEIN PHOSPHATASE 2A holoenzyme assembly and plays important roles in hormone signaling, salt stress response, and plant development. Plant Physiol 166(3): 1519-1534.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chen, J., Shen, G. and Zhang, H. (2015). A Non-Radioactive Method for Measuring PP2A Activity in Plants. Bio-protocol 5(17): e1577. DOI: 10.21769/BioProtoc.1577.
- Chen, J., Hu, R., Zhu, Y., Shen, G. and Zhang, H. (2014). Arabidopsis PHOSPHOTYROSYL PHOSPHATASE ACTIVATOR is essential for PROTEIN PHOSPHATASE 2A holoenzyme assembly and plays important roles in hormone signaling, salt stress response, and plant development. Plant Physiol 166(3): 1519-1534.
Category
Plant Science > Plant biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









