- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Spot Assays for Viability Analysis of Cyanobacteria
Published: Vol 5, Iss 17, Sep 5, 2015 DOI: 10.21769/BioProtoc.1574 Views: 11910
Reviewed by: Maria SinetovaElizabeth LibbyAmit Dey

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of the Predatory Capability of Bdellovibrio bacteriovorus HD100
Cristina Herencias [...] Virginia Martínez
Mar 20, 2017 11623 Views

Determination of Survival of Wildtype and Mutant Escherichia coli in Soil
Yinka Somorin and Conor O'Byrne
Jul 20, 2017 8838 Views

Shipment of Cyanobacteria by Agarose Gel Embedding (SCAGE)—A Novel Method for Simple and Robust Delivery of Cyanobacteria
Phillipp Fink [...] Karl Forchhammer
Dec 5, 2024 1398 Views
Abstract
Cyanobacteria are prokaryotic organisms performing oxygenic photosynthesis. The cyanobacterium Synechocystis sp. PCC 6803 is a model organism for the study of photosynthesis, gene regulation and biotechnological applications because it is easy to manipulate genetically. Moreover, this cyanobacterium can grow photoautotrophically as well as chemoheterotrophically in the dark utilizing glucose. Microbiologists often use optical density measured with a spectrophotometer for the comparison of growth performance of different strains in liquid cultures. Because Synechocystis sp. PCC 6803 (especially motile strains) tend to form aggregates under stress conditions this method might be not suitable for evaluation of different strains under different growth conditions. In addition, many labs are not well equipped with standardized photobioreactors and illumination facilities to ensure reproducibility of growth curves. Here, we describe a highly reproducible spot assay for viability analysis of Cyanobacterial strains.
Keywords: CyanobacteriaMaterials and Reagents
- Synechocystis sp. PCC 6803 strain (wild type obtained from S. Shestakov, Moscow State University, Russia)
- CaCl2.2H2O (Carl Roth, catalog number: 5239.1 )
- Citric acid (Carl Roth, catalog number: X863.2 )
- NaNO3 (Carl Roth, catalog number: A136.2 )
- MgSO4.7H2O (Carl Roth, catalog number: P027.1 )
- Na2-EDTA (Carl Roth, catalog number: 8043.2 )
- H3BO3 (Carl Roth, catalog number: 6943.1 )
- MnCl2.4H2O (Carl Roth, catalog number: T881.3 )
- ZnSO4.7H2O (Carl Roth, catalog number: T884.1 )
- Na2MoO4.2H2O (Carl Roth, catalog number: 0274.1 )
- Co(NO3)2.6H2O (Carl Roth, catalog number: HN16.1 )
- CuSO4.5H2O (Carl Roth, catalog number: P024.1 )
- Na2CO3 (Carl Roth, catalog number: P028.2 )
- Ammonium iron (III) citrate (Carl Roth, catalog number: P027.1)
- K2HPO4.3H2O (Merck KGaA, Calbiochem®, catalog number: 105099 )
- 2-[(2-Hydroxy-1, 1-bis(hydroxymethyl)ethyl)amino]ethanesulfonic acid (TES) (Carl Roth, catalog number: 9137.3 )
- Bacto Agar (BD Bioscience, catalog number: 214010 )
- Syringes (50 ml volume)
- Sterile syringe filters (Sarstedt Filtropur S 0.2 µm, catalog number: 83.1826.001 and 0.45 µm, catalog number: 83.1826 )
- Sterile 96-well microliter plates (i.e. VWR International, catalog number: 734-2328 )
- 100x BG11 medium (see Recipes)
- Trace metal mix (see Recipes)
- Stock Solutions for BG-11 medium (see Recipes)
- 1x BG11 (2x BG11) (see Recipes)
- BG11 agar (see Recipes)
- Na2S2O3 solution (see Recipes)
- 0.25 M Na2-EDTA (see Recipes)
Equipment
- Wide neck Erlenmeyer flasks with 100 ml volume (Carl Roth, catalog number: C146.1 )
- Light source (18 W/840) (Philips, model: MASTER TL-D Super 80 )
- Laboratory shaker (one-dimensional orbital motion) (Heidolph Instruments GmbH, catalog number: 036130180 )
- UV/Vis spectrophotometer (Shimadzu, model: UV-2401PC )
- Multichannel pipettes (8-channel pipette, 0.5-10 µl and 10-100 µl) (Starlab ErgoOne®, catalog number: S7108-0510 and S7108-1100 )
- Petri dishes with square shape (120 x 120 x 17 mm) (Greiner Bio-One GmbH, catalog number: 688102 )
- Transmitted light scanner with adjustable cover (i.e. Epson perfection v700 photo, a good camera can be used as an alternative)
- Laminar flow hood (i.e. Heraeus HeraSafe HS)
- LI-190R Quantum Sensor
Procedure
Every experimental step involving the handling of cyanobacterial cell cultures must be performed under a Laminar flow hood to prevent contamination!
- Cyanobacterial liquid cultures were inoculated from agar plates in BG11 medium (Rippka et al., 1979) and incubated with continuous shaking (150 rpm) at 30 °C under constant illumination with white light of 50 µmol photons m-2 s-1 measured at the position of the agar plates for 2-3 days (measured with a LI-190R Quantum Sensor).
Note: Even though some mutants carry antibiotic resistance gene cassettes inserted in their genome, cultures for the spot assays were always grown in medium lacking antibiotics to exclude effects from growth on selective media. There is no specific volume or OD needed, but the pre-cultures should not reach late stationary phase and their OD750 should be similar.
- The well-grown pre-cultures were diluted to OD750 0.2 in a final volume of 20 ml BG11 medium and were incubated further under the above mentioned conditions until they reached logarithmic phase (approximately OD750 0.6-0.7).
Note: It is important to measure OD at a wavelength where no pigments absorb.
- The logarithmically growing cultures were again diluted to OD750 0.2. With the freshly diluted cultures, a dilution series was performed with a multichannel pipette in a sterile microliter plate ranging from 100 to 10-7 (Figure 1).
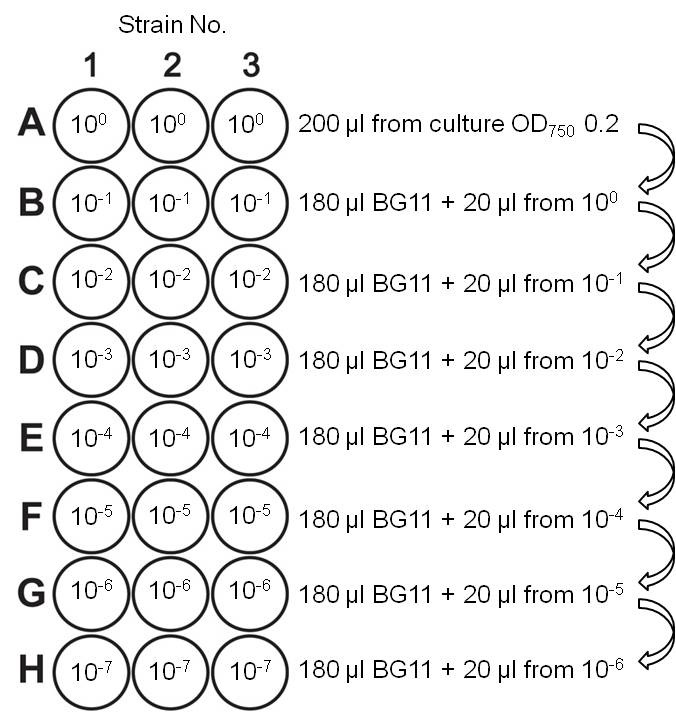
Figure 1. Dilution series for viability analysis. Rows B to H were pre-filled with 180 µl BG11 medium. After every dilution step, the cell suspension was thoroughly mixed with the pipette before transferring part of it into the next well.
- From each of the dilution steps, 5 µl was spotted in triplicates using the multichannel pipette onto square BG11 agar plates supplemented with 0.3% (w/v) Na2S2O3.
Note: The exact composition of the agar can be customized. If the experimenter wishes to observe viability under mixotrophic conditions, the BG11 agar can be i.e. supplemented with glucose. It is also possible to exclude iron or nitrogen from the media stock solutions. However do not forget to add Na2S2O3 to the plates. This is important for proper growth of the colonies (Thiel et al., 1989).
- The drops of liquid cultures were allowed to dry for approximately 15-20 min (closed plate, at room temperature on the table top). Afterwards, plates were incubated at 30 °C under constant illumination with white light at 50 µmol photons m-2 s-1 for approximately 5 days or until no more colonies appeared in the highest dilution step.
Note: To prevent phototactic movement of motile strains, the plates should be placed below the light source and lateral illumination from only one side should be avoided. In order to prevent dripping (in case the drops are not completely dry), the plates were incubated with the lid facing upwards for the first day. After the first day, plates were turned upside down to avoid evaporated water dripping on the spots. For investigation of light-dark related phenotypes, constant illumination can also be changed to light-dark-cycles. In our case, we investigated mutants of circadian clock associated genes and therefore chose a 12:12 h light:dark incubation. In this case, the starting OD750 for the dilution series was adjusted to 0.4 instead of 0.2 (step 2).
- Record the result (Figure 2) by scanning the plate or by taking a picture. In the best case, it may even be possible to count the single colonies within several spots (you may need a Binocular microscope).
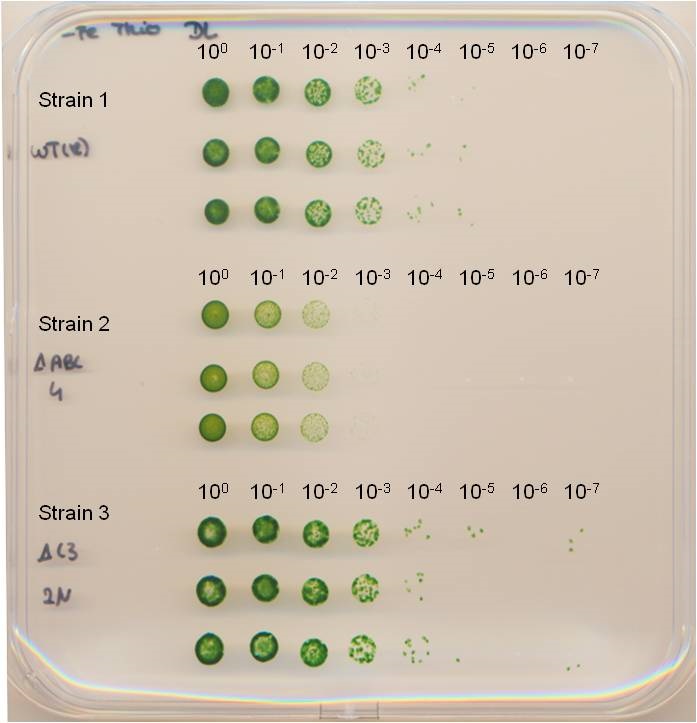
Figure 2. Spot assay. The dilution series of the cell cultures were spotted in triplicates onto square BG11 agar plates. The plate was incubated until no more colonies appeared in the highest dilution step. Strains have been published by Dörrich et al. (2014).
Further results generated by using this method were published by Dörrich et al. (2014).
Notes
The spot assays described above are highly reproducible. In contrast to other methods like measuring the optical density of liquid cultures, which is often error-prone due to i.e. the formation of aggregates, we always obtained reliable and reproducible results. Within our group, several researchers have performed this method in two different laboratories and we did not detect any variations in the results. For reproducibility it is highly recommended to use Na2S2O3 in the plates (as described in Recipes) and to record the results after the same time. Composition of agar plates and light conditions can be customized to fit the experimenters’ individual needs and may require optimization for different mutant strains.
Recipes
- 100x BG11 medium
Prepare the solution in ddH2O and sterilize by autoclavingCaCl2.2H2O
3.6 g/L
Citric acid
0.6 g/L
NaNO3
149.58 g/L
MgSO4.7H2O
7.49 g/L
0.25 M Na2-EDTA (pH 8)
0.56 ml/L
- Trace metal mix
Prepare these solutions in ddH2O and sterilize by filtering using a syringe and sterile filterH3BO3
2.86 g/L
MnCl2.4H2O
1.81 g/L
ZnSO4.7H2O
0.222 g/L
Na2MoO4.2H2O
0.390 g/L
Co(NO3)2.6H2O
0.049 g/L
Sterile aliquots can be stored at 4 °C
- Stock Solutions for BG-11 medium
Prepare these solutions in ddH2O and sterilize by filtering using a syringe and sterile filterNa2CO3
20 mg/ml
Ammonium iron (III) citrate
6 mg/ml
K2HPO4.3H2O
30 mg/ml
TES buffer (pH 8.0 adjust with NaOH)
1 M
Sterile aliquots can be stored at 4 °C
- 1x BG11 (2x BG11)
Add sterile solutions to ddH2O and adjust the volume to 1 L100 x BG11
10 ml/L (20 ml/L)
Na2CO3 stock solution
1 ml/L (2 ml/L)
K2HPO4.3H2O stock solution
1 ml/L (2 ml/L)
Ammonium iron (III) citrate stock solution
1 ml/L (2 ml/L)
Trace metal mix stock solution
1 ml/L (2 ml/L)
TES buffer (pH 8.0) stock solution
10 ml/L (20 ml/L)
If you use non-sterile solutions and autoclave the medium afterwards there will be slight precipitation of salts. Before using the medium you should mix it thoroughly. This is recommended, when you want to avoid contaminations in your cultures, especially for long-term strain maintenance.
- BG11 agar
Prepare 1.5% bactoagar in ddH2O and autoclave.
The molten 1.5% bactoagar (approx. 80 °C) is mixed with the same volume of sterile 2x BG11 medium.
- Na2S2O3 solution
30% w/v Na2S2O3
30 g Na2S2O3
Add ddH2O to 100 ml
Filter sterilize (0.2 µm)
Stored at 4 °C
- 0.25 M Na2-EDTA (pH 8)
9.306 g Na2-EDTA
Add ddH2O to 100 ml
Adjust pH to 8.0 with NaOH (use highly concentrated NaOH, Na2-EDTA will not dissolve, until the pH reaches 8.0)
Stored at room temperature
Acknowledgments
BG11 medium is prepared according to Rippka et al. (1979). This work was supported by DFG grant to A. W. Wi2014/5-1.
References
- Dörrich, A. K., Mitschke, J., Siadat, O. and Wilde, A. (2014). Deletion of the Synechocystis sp. PCC 6803 kaiAB1C1 gene cluster causes impaired cell growth under light-dark conditions. Microbiology 160(Pt 11): 2538-2550.
- Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M. and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. J Gen Microbiol (111): 1-61.
- Thiel, T., Bramble, J. and Rogers, S. (1989). Optimum conditions for growth of Cyanobacteria on solid media. FEMS Microbiol Lett 52(1-2): 27-31.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Dörrich, A. K. and Wilde, A. (2015). Spot Assays for Viability Analysis of Cyanobacteria. Bio-protocol 5(17): e1574. DOI: 10.21769/BioProtoc.1574.
Category
Microbiology > Microbial cell biology > Cell viability
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









