- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Immunolocalization of Proteins in Corals: the V-type H+-ATPase Proton Pump
Published: Vol 5, Iss 17, Sep 5, 2015 DOI: 10.21769/BioProtoc.1573 Views: 11077
Reviewed by: Arsalan DaudiClaudia CatalanottiModesto Redrejo-Rodriguez

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Non-separate Mouse Sclerochoroid/RPE/Retina Staining and Whole Mount for the Integral Observation of Subretinal Layer
Sung-Jin Lee and Soo-Young Kim
Jan 5, 2021 4457 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views

Time-Lapse Into Immunofluorescence Imaging Using a Gridded Dish
Nick Lang [...] Andrew D. Stephens
Feb 20, 2026 48 Views
Abstract
Here we describe the immunolocalization of a membrane-bound proton pump, the V-type H+-ATPase (VHA), in tissues and isolated cells of scleractinian corals. Immunolocalization of coral proteins requires additional steps not required for various model organisms, such as decalcification of the coral skeleton for immunohistochemistry or removal of cells away from the skeleton for immunocytochemistry. The tissue and cell preparation techniques described here can be adapted for localization of other coral proteins, provided the appropriate validation steps have been taken for the primary antibodies and species of coral used. These techniques are important for improving our understanding of coral cell physiology
Materials and Reagents
- Live coral
- Electron microscopy grade paraformaldehyde (16% solution in 10 ml ampules) (Electron Microscopy Sciences, catalog number: 15700 or 15710 )
- Ethyl alcohol (absolute, 200 proof)
- Ethyl alcohol (190 proof)
- Xylenes (any reagent ≥ 98.5% xylenes, other tissue clearing agents such as Safeclear, are also acceptable) (Safeclear, catalog number: 044-192 )
- Paraffin embedding medium (e.g. Paraplast X-TRA) (McCormick Scientific, catalog number: 39503002 )
- Triton-X-100 (Bio-Rad Laboratories, AbD Serotec®, catalog number: 1610407 or Sigma-Aldrich, catalog number: T8787 )
- Normal goat serum (Vector laboratories, catalog number: S-1000 )
- Keyhole Limpet Hemocyanin (Sigma-Aldrich, catalog number: 7017 )
- Phosphate buffered saline (PBS) (e.g. 10x) (Corning Incorporated, catalog number: 46-013-CM )
- Microscope slides and coverslips
- Custom, antigen-affinity purified, rabbit polyclonal anti-VHAB antibodies developed using a peptide antigen matching a conserved region of the VHAB subunit (AREEVPGRRGFPGY) (Genscript USA, Inc.). The epitope is 100% conserved among diverse animal species, and specifically recognizes VHAB from coral (Barott et al., 2015), bone-eating worms (Tresguerres et al., 2013), hagfish (Clifford, et al. 2015), and sharks (Tresguerres et al., 2010; Roa et al., 2014).
Note: This protocol may be adapted for other proteins following validation of the primary antibodies in the coral species to be examined. However, successful localization using different primary antibodies may require optimization of the fixation procedure from that described below. - Secondary antibodies: Goat anti-rabbit IgG Alexa Fluor 555 (2 mg/ml) (Life Technologies, catalog number: A21429 )
Note: The Alexa555 fluorophore is used to avoid overlap with the coral’s endogenous green fluorescent protein (GFP), but other fluorophores outside of the GFP emission range may also work. - Hoescht 33342 (Life Technologies, catalog number: H1399 )
- Mounting medium (e.g. Electron Microscopy Sciences, catalog number: 17895 )
Note: Unless otherwise noted, reagents can be obtained from any general laboratory reagent manufacturer or supplier. - S22 buffer (see Recipes)
- Ca-free S22 buffer + 0.5 M EDTA (see Recipes)
- Fixative solution (see Recipes)
- Blocking buffer (see Recipes)
Equipment
- Chemical fume hood
- Fluorescence microscope (e.g. Zeiss AxioObserver Z1, or any other microscope with the ability to detect blue (“DAPI filter”, peak excitation 350 nm; peak emission 461 nm; for Hoescht staining of nuclei) and red (“TRITC filter”, peak excitation 550 nm; peak emission 570 nm; for secondary antibodies) fluorescence.
- Optional
- Coral endogenous GFP and chlorophyll from dinoflagellate symbionts (Symbiodinum) are visible using settings for “green fluorescence” (“FITC filter”, peak excitation 490 nm; peak emission 525 nm).
- Structured illumination (Zeiss Apotome2), confocal capabilities *suggested but not required.
- Differential Interference Contrast (DIC) (Nomarski microscopy) *suggested but not required.
- Coral endogenous GFP and chlorophyll from dinoflagellate symbionts (Symbiodinum) are visible using settings for “green fluorescence” (“FITC filter”, peak excitation 490 nm; peak emission 525 nm).
- Microcentrifuge [any model capable of spinning 1.5 ml tubes at 3,000 x g at 4 ℃
Note: This temperature can be achieved by placing the microcentrifuge inside of a fridge. - Tissue cassettes (e.g. C&A Scientific, catalog number: EC-0109x )
- Tissue embedding station or any other method to pour liquid paraffin (e.g. Kedee, model: KD-BM II )
- Embedding base molds (metal) (e.g. C&A Scientific, catalog number: HB-07 , 15 , 24 , 30 )
- Embedding rings (plastic) (e.g. Biologix, catalog number: 41-3004 )
- Microtome (any model capable of cutting 5-30 μm paraffin sections)
- Slide warmer/hot plate (any model capable of warming up slides to 30 ℃)
- Hydrobarrier pen (Pap pen) (e.g. Vector Laboratories, catalog number: H-4000 )
- Rotating platform (any rotator/shaker/rocker)
Software
- Imaging software (e.g. Zeiss Axiovision software, Adobe Photoshop, ImageJ)
Procedure
- Tissue preparation
Note: Section A takes approximately 10-14 days when completed without stopping. There are several stopping places within Section A.- Immerse a live coral fragment (~1-2 cm in length) in fixative solution [S22 buffer with 3% paraformaldehyde (PFA), see Recipes] and incubate overnight at 4 °C (Puverel et al., 2005).
Notes:- PFA stock solution should be handled in a chemical fume hood.
- It is best to add PFA stock solution (16% or 32% PFA) to S22 buffer immediately before use. Diluted PFA can be stored at 4 °C for up to one week. Stock PFA may be stored at 4 °C for up to one month or at -20 °C for 6 months once unsealed.
- PFA stock solution should be handled in a chemical fume hood.
- Decalcify by transferring fragment to Ca-free S22 buffer with 0.5 M EDTA and 0.5% paraformaldehyde and incubate at 4 °C. Replace the buffer daily until the skeleton is completely dissolved (~7 days depending on size of fragment and thickness/density of skeleton). Dissolution of the skeleton is determined visually; the coral tissue should appear transparent with no visible white calcium carbonate remaining (Figure 1).
Note: It is best to add PFA stock solution (16% or 32% PFA) to Ca-free S22 buffer immediately before use.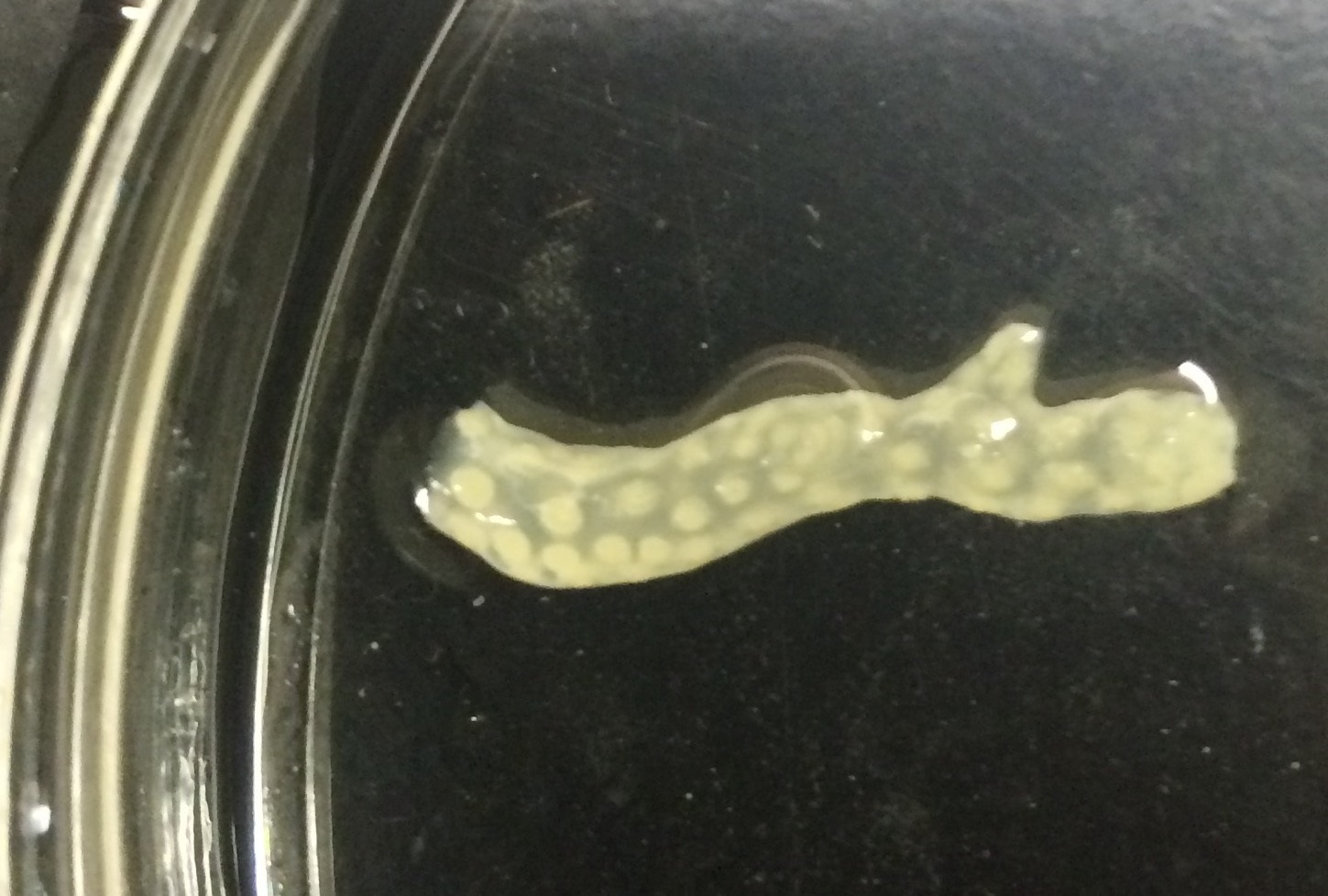
Figure 1. Example of a fully decalcified coral fragment. Only the tissue remains. - Dehydrate tissue.
Note: Incubations in steps a-f are conducted on a shaking platform.- Incubate in 50% ethanol for 2-5 h.
- Incubate in 70% ethanol overnight at 4 °C.
- Tissue will keep in 70% ethanol at 4 °C for months.
- Put tissue in mesh plastic tissue cassette for final incubation steps in glass beaker.
- Incubate in 95% ethanol for 20 min.
- Incubate in 100% ethanol for 20 min. Repeat with fresh ethanol for a total of three times.
- Incubate in xylene for 20 min. Repeat with fresh xylene for a total of three times.
- **Do in HOOD.
- Incubate in paraffin for 30 min. Repeat with fresh paraffin for a total of three times.
- Incubate in 50% ethanol for 2-5 h.
- Embed tissue in paraffin wax.
- Transfer tissue to base mold and position as desired.
- Fill with wax.
- Place embedding ring on top of base mold, and fill with wax to the top.
- Let wax solidify at room temperature for 1-2 days.
- Embedded tissue may be stored at room temperature* for several months prior to sectioning; long-term, archival storage must be done at 4 °C (*but storage at 4 °C is recommended).
- Transfer tissue to base mold and position as desired.
- Sectioning.
- Turn on hot plate to 40 °C.
- Trim wax block around the coral tissue into the shape of a trapezoid (i.e. so top and bottom are parallel, bottom edge longer, and left half square; Figure 2).
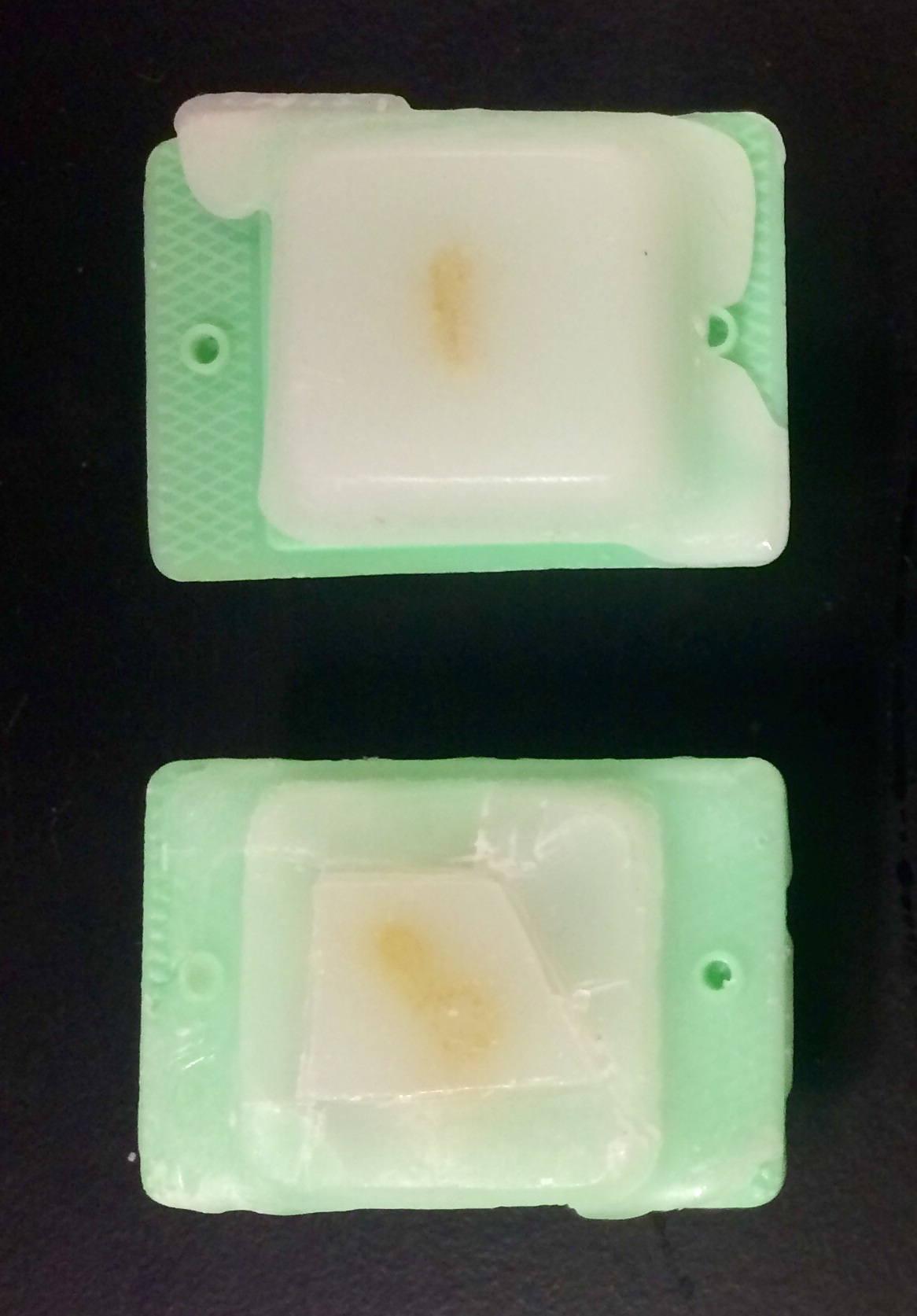
Figure 2. Example of coral tissue embedded in paraffin wax before (top) and after trimmed (bottom) in preparation for sectioning by a microtome - Load sample onto microtome.
- Load blade with holder.
- Blade stored at -20 °C.
- Blade set at 15 degree angle (may be adjusted).
- Blade stored at -20 °C.
- Unlock stage and slide until blade almost touching sample.
- Start cutting sample using thick (15-30 μm) sections until surface is flat and smooth and tissue is reached.
- Switch to desired tissue thickness for slides (5-10 μm).
- Should get ribbon of consecutive sections (each section is easily identifiable because the right edge of each section is not parallel to the left edge of the following section).
- Cut ribbon into individual sections with razor blade and place onto glass microscope slides.
- Blade-cut side down.
- Label slides with pencil.
- Blade-cut side down.
- Float section on water, remove any bubbles (two or three consecutive sections can be placed on each slide).
- Stretch wax section by placing slide on hot plate (~30 sec). Surface should become smooth.
- Remove water with Kim wipe.
- Let dry/affix to slide by leaving slide on hotplate overnight at 30 °C with paper towel underneath.
- Store at 4 °C until use.
- Turn on hot plate to 40 °C.
- Deparaffinization and rehydration.
- Incubate in xylene bath for 10 min. Repeat with fresh xylene for a total of three times.
- **Do in HOOD.
- Incubate in 100% ethanol bath for 10 min.
- Incubate in 95% ethanol bath for 10 min.
- Incubate in 70% ethanol bath for 10 min.
- Incubate in PBS-TX (1x PBS with 0.2% (v/v) Triton-X-100) bath for 10 min.
- Incubate in xylene bath for 10 min. Repeat with fresh xylene for a total of three times.
- Isolated coral cells were prepared from A. yongei colonies as described previously (Venn et al., 2009; Barott et al., 2015). Briefly, a coral fragment was submerged in 0.2 μm filtered seawater and the tissue was removed from the skeleton by gentle brushing with a toothbrush. Cells were then pelleted by centrifugation (3,000 x g for 4 min at 4 °C) and resuspended in the desired solution (see step B2 below).
- Fix cells by incubating in S22 buffer with 4% paraformaldehyde for 15 min on ice.
- Pellet cells (3,000 x g for 4 min at 4 °C) to remove the fixative. Resuspend and concentrate cells in 500 µl S22 buffer.
- Spread onto glass slides and air-dry. Optional: This step may be repeated several times to increase the cell concentration on each slide.
- Stain sample right away as described in section C below; if absolutely necessary slides may be stored at 4 °C for up to 1 h prior to staining.
C.Staining- Draw circle around tissue with hydrobarrier pen (Figure 3).
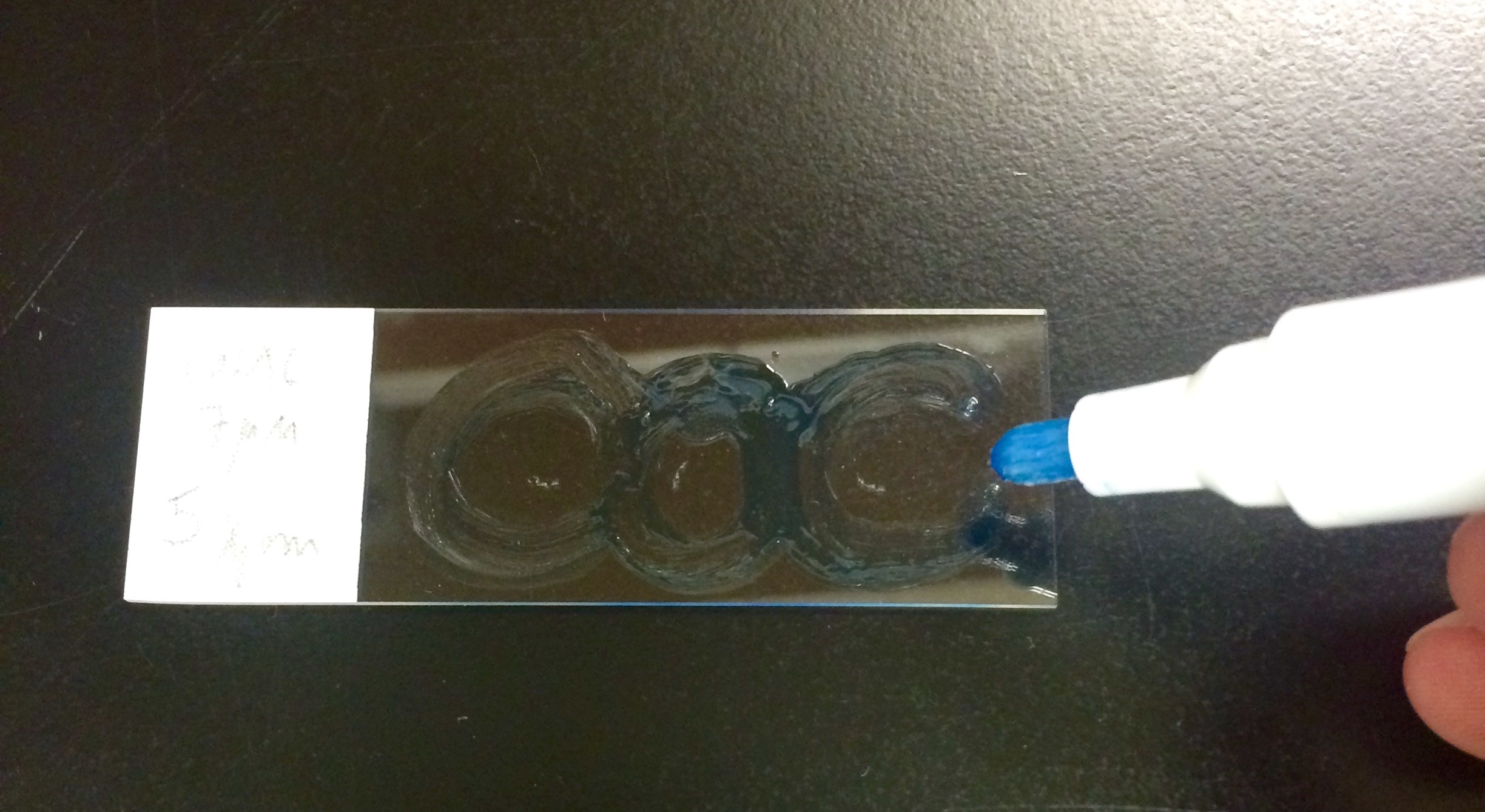
Figure 3. Example of slide containing three tissue sections encircled by hydrobarrier - Permeabilize tissue sections or isolated cells by adding PBS-TX to the top of the section/cells (~70-100 µl).
- Incubate for 60 or 10 min, respectively.
- Remove liquid using pipet. Incubate samples with 70-100 µl blocking buffer for 1 h at room temperature.
- Incubate samples with the anti-VHAB antibodies (3 µg/ml in blocking buffer) overnight at 4 °C in a humidified chamber.
- For secondary antibody controls, blocking buffer with no primary antibody is used on a section on the same slide.
Note: Antibody specificity must be confirmed for each new species and antibodies pool; see below. - Rinse in sequential baths of PBS-TX for 5 min each (3 times total).
- Incubate with the secondary antibody (goat anti-rabbit-Alexa 555; 1:500 dilution in blocking buffer) for 1 h at room temperature (secondary antibodies with other fluorophores may also be used; however, many coral cells, especially those in the polyp tentacles, contain very bright green fluorescent green protein that masks fluorophores such as Alexa 488 and similar).
- Incubate with Hoescht (1 µg/ml) for 5 min at room temperature to stain DNA.
- Rinse in sequential baths of PBS-TX for 5 min each (3 times total).
- Add ~50 µl mounting gel to top of each section.
- Place glass cover slip on top.
- Smooth away bubbles from tissue region by sliding the coverslip around (do not pick up cover as it may damage the tissue).
- Place glass cover slip on top.
- Dab edges of cover slip with nail polish to secure, allow to dry.
- Store at 4 °C until imaging.
D.Validation of anti-VHAB antibodies specificity
Note: Validation of antibodies by Western blot should be done on homogenized samples. A single, clean band of approximately 55 KDa should be obtained for anti-VHAB antibodies, this band should not be present in peptide pre-absorption control, pre-immune serum control, or when the primary antibodies are omitted. Roa et al. (2014) describes peptide pre-absorption controls in detail, and Barott et al. (2015) describes preparation of coral samples for polyacrylamide gel electrophoresis and Western blot. These same controls should be carried out for IHC, and should lead to low background signal (see example in Figure 4).- Prepare the following solutions in blocking buffer:
- Pre-immune serum control (3 µg/ml).
- Peptide pre-absorption control (3 µg/ml anti-VHAB antibodies plus 6 µg/ml peptide antigen). Incubate in rotating platform overnight at 4 °C.
- Anti-VHAB antibodies (3 µg/ml). Incubate in rotating platform overnight at 4 °C.
- Pre-immune serum control (3 µg/ml).
- Incubate samples as described in “Procedure C”. Staining, steps C4-10.
E.Microscopy- Observe and capture images using standard immunofluorescence microscopy protocols (epifluorescence, structured illumination, confocal), and appropriate excitation/emission settings for each fluorophore.
- If using Alexa555 secondary antibodies, VHA immunofluorescence is visible in the red channel (Figure 4). At longer exposure times, chlorophyll autofluorescence from dinoflagellate symbionts (Symbiodinum), will also be visible.
- Adjust the settings to the longest exposure time that results in maximum VHA signal from host coral cells and minimum or no chlorophyll signal. Control sections should show no red fluorescence under using the same settings.
- Visualize nuclei using blue fluorescence channel settings. Nuclei in coral host cells appear as solid circles, while nuclei in symbionts generally display a punctate staining pattern (possibly caused by super-coiled chromosomes).
- Optional 1: Visualize coral endogenous green fluorescence proteins and symbiont chlorophyll autofluorescence using green fluorescence channel settings.
- Optional 2: Visualize general tissue morphology using DIC.
- Use imaging software to adjust digitalized images for brightness and contrast only over the entire image (and following standard image manipulation guidelines).
- Immerse a live coral fragment (~1-2 cm in length) in fixative solution [S22 buffer with 3% paraformaldehyde (PFA), see Recipes] and incubate overnight at 4 °C (Puverel et al., 2005).
Representative data
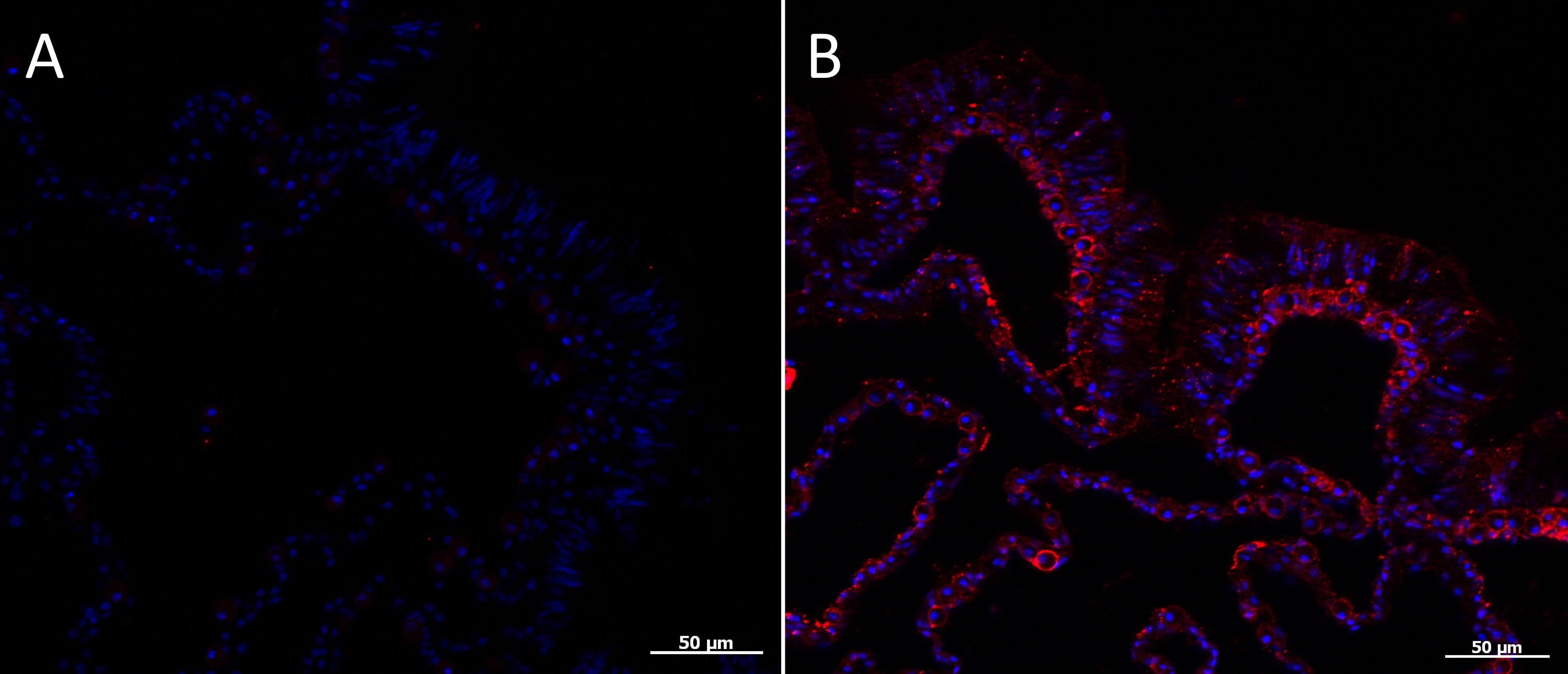
Figure 4. Immunofluorescence localization of VHA in tissues of the coral Acropora yongei. A. Control (primary antibodies omitted), B. anti-VHAB antibodies (red). Hoescht staining of nuclei is shown in blue.
Recipes
- S22 buffer
- 450 mM NaCl
- 10 mM KCl
- 58 mM MgCl2
- 10 mM CaCl2
- 100 mM Hepes (pH 7.8)
Note: Adjust pH of Hepes buffer with NaOH.
- 450 mM NaCl
- Ca-free S22 buffer + 0.5 M EDTA
- a450 mM NaCl
- 10 mM KCl
- 58 mM MgCl2
- 0.5 M EDTA
- 100 mM Hepes (pH 7.8)
Note: Adjust pH of Hepes buffer with NaOH.
- a450 mM NaCl
- Fixative solution (10 ml)
- Break open PFA ampule (16% PFA stock solution).
- Add 1.9 ml PFA stock solution to 8.1 ml S22 buffer (3% PFA final concentration). Invert to mix.
Notes:- Remaining PFA stock solution can be stored at 4 °C for up to one month or at -20 °C for 6 months.
- Fixative solution can be stored at 4 °C for up to one week.
- Remaining PFA stock solution can be stored at 4 °C for up to one month or at -20 °C for 6 months.
- Break open PFA ampule (16% PFA stock solution).
- Blocking buffer
- 4 ml PBS-0.2% TritonX
- 80 µl NGS (normal goat serum)
- 0.8 µl KLH (Keyhole Limpet Hemocyanin) solution
- 4 ml PBS-0.2% TritonX
Acknowledgments
This work was supported by National Science Foundation Grants EF-1220641 (to M. T.) and OCE-1226396 (to K. L. B.), and by Alfred P. Sloan Research Fellowship Grant BR2013-103 (to M. T.). We thank Sidney Perez for assistance with the isolated cell staining protocol.
References
- Barott, K. L., Venn, A. A., Perez, S. O., Tambutte, S. and Tresguerres, M. (2015). Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. Proc Natl Acad Sci U S A 112(2): 607-612.
- Clifford, A., Goss, G. G., Roa, J. N. and Tresguerres, M. (2015). in Hagfish Biology [Science Publishers, CRC Press, Boca Raton, FL (In press)].
- Puverel, S., Tambutté, E., Zoccola, D., Domart-Coulon, I., Bouchot, A., Lotto, S., Allemand, D. and Tambutté, S. (2005). Antibodies against the organic matrix in scleractinians: a new tool to study coral biomineralization. Coral Reefs 24(1): 149-156.
- Roa, J. N., Munevar, C. L. and Tresguerres, M. (2014). Feeding induces translocation of vacuolar proton ATPase and pendrin to the membrane of leopard shark (Triakis semifasciata) mitochondrion-rich gill cells. Comp Biochem Physiol A Mol Integr Physiol 174: 29-37.
- Tresguerres, M., Katz, S. and Rouse, G. W. (2013). How to get into bones: proton pump and carbonic anhydrase in Osedax boneworms. Proc Biol Sci 280(1761): 20130625.
- Tresguerres, M., Parks, S. K., Salazar, E., Levin, L. R., Goss, G. G. and Buck, J. (2010). Bicarbonate-sensing soluble adenylyl cyclase is an essential sensor for acid/base homeostasis. Proc Natl Acad Sci U S A 107(1): 442-447.
- Venn, A. A., Tambutte, E., Lotto, S., Zoccola, D., Allemand, D. and Tambutte, S. (2009). Imaging intracellular pH in a reef coral and symbiotic anemone. Proc Natl Acad Sci U S A 106(39): 16574-16579.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Barott, K. L. and Tresguerres, M. (2015). Immunolocalization of Proteins in Corals: the V-type H+-ATPase Proton Pump. Bio-protocol 5(17): e1573. DOI: 10.21769/BioProtoc.1573.
Category
Biochemistry > Protein > Immunodetection > Immunostaining
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










