- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Separation of Intracellular Vesicles for Immunoassays
Published: Vol 5, Iss 16, Aug 20, 2015 DOI: 10.21769/BioProtoc.1571 Views: 12630
Reviewed by: Jia LiHsin-Yi ChangLee-Hwa Tai

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Generation of T cells from Human and Nonhuman Primate Pluripotent Stem Cells
Akhilesh Kumar [...] Igor I. Slukvin
Jul 5, 2020 7286 Views

Expansion and Polarization of Human γδT17 Cells in vitro from Peripheral Blood Mononuclear Cells
Xu Chen [...] Jun Yan
Jan 5, 2024 1951 Views

Proliferation Assay Using Cryopreserved Porcine Peripheral Mononuclear Cells Stimulated With Concanavalin A and Analyzed With FCS ExpressTM 7.18 Software
Marlene Bravo-Parra [...] Luis G. Giménez-Lirola
Jun 5, 2025 2620 Views
Abstract
The endosome/lysosome systems play important roles in immune cell functions as signaling platforms. Immune cells utilize these endosome/lysosome for signal transduction or intercellular communication to elicit the proper immune responses, regulating the localization or the association of the signaling complexes. Here we introduce the procedures to separate the intracellular vesicles such as endosomes or lysosomes, which could be useful to identify the subcellular localization of the signaling complexes.
Materials and Reagents
- Cells (e.g. RAW264.7, Raji, mouse naïve B cells)
- Trypan blue staining (0.4% solution) (Life Technologies, catalog number: 15250-061 )
- CD43-microbeads (Miltenyi Biotec, catalog number: 130-049-801 )
- Blocking One (Nacalai tesque, catalog number: 03953-95 )
- Anti-mouse LAMP1 (eBioscience, catalog number: 1D4B )
- Anti-GM130 (BD biosciences, catalog number: 35/GM130 )
- Anti-Rab5 (Santa Cruz Biotechnologies, catalog number: D-11 )
- Anti-Calnexin (Enzo Life Sciences, catalog number: ADI-SPA-860 )
- Anti-COX IV (Cell Signaling Technology, catalog number: 4844 )
- Anti-rat Ig(H+L)-HRP (Jackson ImmunoResearch Laboratories, catalog number: 712-036-153 )
- Anti-mouse Ig(H+L)-HRP (Jackson ImmunoResearch Laboratories, catalog number: 115-035-062 )
- Anti-rabbit Ig(H+L)-HRP (Jackson ImmunoResearch Laboratories, catalog number: 111-035-144 )
- ECL substrate (Thermo Fisher Scientific, catalog number: 34095 )
- Protease Inhibitor Cocktail (Thermo Fisher Scientific, catalog number: 87785 )
- Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, catalog number: 78420 )
- 60% OptiPrep (Axis-Shield plc, catalog number: 1114542 )
- Solution A (see Recipes)
- Solution B (see Recipes)
- Solution C (see Recipes)
- Solution D (see Recipes)
- Solution E (see Recipes)
- 30%, 23%, 17%, 11%, and 5% Optiprep (2 ml each) (see Recipes)
- TBS-T (see Recipes)
Equipment
- 10 cm dish
- Centrifuge tube (Beckman Coulter, catalog number: 347357 )
- Hemocytometer
- Centrifuge (with cooling function will be recommended)
- 1 ml syringe with 29-gauge needle
Procedure
Protocol A should be chosen when regular cell-line (e.g. RAW264.7, Raji, or HEK293T) or cells having large cytosol in volume are analyzed whereas Protocol B should be chosen for small cells or cells having small cytosol, such as lymphocytes.
- Procedures for cell extraction from 10 cm dish (2 dishes for each sample)
- Layer 360 µl of the each gradient solution (30%, 23%, 17%, 11%, and 5% OptiPrep) in the Centrifuge tube(s) with P200 micropipette, as shown in Figure 1.

Figure 1. Illustration showing the way to layer the gradient solution using P200 micropipette. Pour 360 µl of each gradient solution carefully into a centrifuge tube using P200 micropipette (180 µl x 2 times), not to make bubbles nor disturb the other layers. Layer in a centrifuge tube from the heaviest (30%) to lightest (5%) solution. - Leave tubes at 4 °C for more than one hour and keep it still until use.
- Confirm that cells (e.g. RAW264.7, or Raji) are healthy and 75-80% confluent under a microscopy.
- (Optional) Treat with stimulants for the desired period (e.g. LPS at 100 ng/ml for 30 min).
- Wash twice with 10 ml of ice-cold PBS, transfer to suitable tubes, and determine cell density and viability with a hemacytometer and trypan blue staining.
- Transfer 1.5 ~ 2.0 x 107 cells to new tubes and centrifuge at 1,100 rpm for 5 min at 4 °C.
- Aspirate the supernatant, tap tubes to loosen cells, and add 450 µl of ice-cold Solution E per tube.
- Homogenize cells by 80-100 passages through a 29-gauge needle until cells show roughly 70-80% trypan blue positive. Cells should be kept cold.
- Centrifuge the homogenate at 1,000 x g for 5 min at 4 °C and transfer the whole supernatant (the post-nuclear fraction) to new 1.5 ml tubes and centrifuge at 1,000 x g for 5 min at 4 °C again. Repeat this step until pellet of cell debris can not be seen after centrifugation (usually 3 times).
Note: The whole supernatant should be taken for the second centrifugation, and the same for the third centrifugation. Initial volume of supernatant after centrifugation should be roughly 400 µl, and after 2 times centrifugation it will be reduced to ca. 350 µl. - Carefully layer 300 µl of the supernatant on the top of the gradient solution made in step A1 (Figure 3).
- Set the centrifuge tubes in the chilled ultracentrifuge rotor TLS-55.
- Centrifuge for 4 h at 39,000 rpm (130,000 x g) at 4 °C by Optima MAX-TL.
Note: Set acceleration (Accel) and deceleration (Decel) at 8 and 0 (free deceleration), respectively. - Harvest 185 µl per one fraction to make 11 fractions using P200 micropipette, and transfer to 1.5 ml microtubes.
- Store at -80 °C until use.
- Layer 360 µl of the each gradient solution (30%, 23%, 17%, 11%, and 5% OptiPrep) in the Centrifuge tube(s) with P200 micropipette, as shown in Figure 1.
- Procedures for mouse naïve B cells
- Layer 360 µl of the each gradient solution (30%, 23%, 17%, 11%, and 5% OptiPrep) in the Centrifuge tube(s) with P200 micropipette.
- Leave tubes at 4 °C for more than one hour and keep it still until use.
Separate the naïve splenic B cells by using CD43-microbeads and MACS according to the manufacturer’s instruction (please see the following link: http://www.miltenyibiotec.co.jp/~/media/Images/Products/Import/0001200/IM0001263.ashx). - (Optional) Treat 1.5 ~ 2.0 x 108 of B cells with stimulants for the desired period (e.g. CpG-1668 at 100 nM for 1 h).
- Wash twice with 10 ml of ice-cold PBS in suitable tubes, and determine cell density and viability with a hemocytometer and trypan blue staining.
- Transfer the cells to new tubes and centrifuge at 1,100 rpm for 5 min at 4 °C.
- Aspirate the supernatant, tap tubes to loosen cells, and add 450 µl of ice-cold Solution E per tube.
- Homogenize cells by passing roughly 200 times through a 29-gauge needle until cells show 70-80% trypan blue positive. Cells should be kept cold on ice.
- Centrifuge the homogenate at 1,000 x g for 5 min at 4 °C and transfer the supernatant (the post-nuclear fraction) to new 1.5 ml tubes. Repeat this step three times.
- Carefully layer 300 µl of the supernatant on the top of the gradient.
- Set the tubes in the chilled ultracentrifuge rotor TLS-55.
- Centrifuge for 4 h at 39,000 rpm (130,000 x g) at 4 °C by Optima MAX-TL.
Note: Set acceleration (Accel) and deceleration (Decel) at 8 and 0 (free deceleration), respectively. - Harvest 290 µl per one fraction to make 7 fractions using P200 micropipette, and transfer to 1.5 ml microtubes.
- Store at -80 °C until use.
- Layer 360 µl of the each gradient solution (30%, 23%, 17%, 11%, and 5% OptiPrep) in the Centrifuge tube(s) with P200 micropipette.
- Evaluation of the fractionation
- Take 30 µl of the fractions and mix with the equal volume of 2x SDS-PAGE sample buffer.
- After brief denaturing at 95 °C, apply 15 µl of the samples to run SDS-PAGE and transfer to PVDF membrane.
- Block the blot with Blocking One, followed by incubation with the primary antibodies against organelle markers. Following antibodies used in this protocol; anti-LAMP1 (dilution 1:1,000) for the detection of the late endosome/lysosome, anti-GM130 for cis-Golgi, anti-Rab5 for endosome, anti-Calnexin for ER, and anti-COX IV for mitochondria, respectively.
- Wash the blots 3 times with TBS-T to remove excess antibodies (5 min x 3 times).
- Incubate with the proper secondary antibodies conjugated with HRP for 1 h [Anti-rat Ig (H+L) for anti-LAMP1, anti-mouse Ig (H+L) for anti-GM130 or anti-Rab5, anti-rabbit Ig (H+L) for anti-Calnexin or COX IV, respectively. Concentration of secondary antibodies is roughly at 50 ng/ml].
- Wash the blots with TBS-T to remove excess antibodies (15 min x 3 times).
- Incubate with ECL substrate, and detect the luminescence signals (Figure 2).
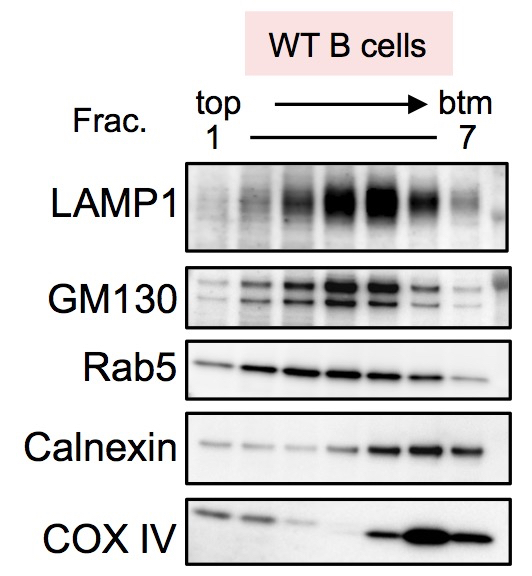
Figure 2. Intracellular vesicles of mouse naïve B cells were fractionated and their distribution was analyzed by western blotting with the specific antibodies against the following organelle markers; late endosome/lysosome with LAMP1, cis-Golgi with GM130, pan-endosome with Rab5, ER with Calnexin, and mitochondria with COX IV, respectively.
- Take 30 µl of the fractions and mix with the equal volume of 2x SDS-PAGE sample buffer.
Notes
- For homogenization of cells, Dounce homogenizer or other devices can be used, as the degree of homogenization difficulty depends on the cell types.
- For small cells such as mouse naïve B cells, number of passage with 29G syringe may differ depending on the cell density.
Recipes
- Solution A
60% OptiPrep - Solution B
OptiPrep diluentAdjust to pH 7.0 with 1 M KOH and make up to 100 ml(Components) 100 ml (Final conc.) H2O ~ 42.8 ml 1 M Hepes (pH7.5) 15 ml 150 mM 1 M KCl 23.5 ml 235 mM 1 M MgCl2 1.2 ml 12 mM 1 M CaCl2 2.5 ml 25 mM 0.2 M EGTA 15 ml 30 mM - Solution C
40% OptiPrep working solution
2 vol. of Solution A + 1 vol. of Solution B - Solution D
Working solution diluent
(Components)100 ml(Final conc.)Adjust to pH 7.0 with 1 M KOH and make up to 100 ml(Components) 100 ml (Final conc.) H2O ~ 81 ml 1 M Hepes (pH7.5) 5 ml 50 mM 1 M KCl 7.8 ml 78 mM 1 M MgCl2 400 µl 4 mM 1 M CaCl2 840 µl 8.4 mM 0.2 M EGTA 5 ml 10 mM - Solution E
Homogenization mediumAdjust to pH 7.0 with 1 M KOH and make up to 100 ml(Components) 100 ml (Final conc.) H2O ~ 81 ml 1 M Hepes (pH7.5) 5 ml 50 mM 1 M KCl 7.8 ml 78 mM 1 M MgCl2 400 µl 4 mM 1 M CaCl2 840 µl 8.4 mM 0.2 M EGTA 5 ml 10 mM Sucrose (FW 342.30) 8.56 g 250 mM
Add Halt Protease Inhibitor Cocktail and Halt Phosphatase Inhibitor Cocktail before use - 30%, 23%, 17%, 11%, and 5% Optiprep (2 ml each)* Halt Protease Inhibitor Cocktail
(Components) 30% 23% 17% 11% 5% Solution C (40% Optiprep) 1500 µl 1150 µl 850 µl 550 µl 250 µl Solution D 500 µl 850 µl 1150 µl 1450 µl 1750 µl Halt Protease Inhibitor* 20 µl 20 µl 20 µl 20 µl 20 µl Halt Phosphatase Inhibitor† 20 µl 20 µl 20 µl 20 µl 20 µl
† Halt Phosphatase Inhibitor Cocktail
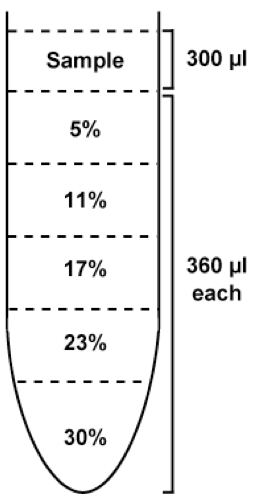
Figure 3. Diagram of completed Opti-prep gradient before ultracentrifugation
- TBS-T
(Components) 1 L (Final conc.) 1 M Tris-HCl (pH7.5) 20 ml 20 mM NaCl 8 g 137 mM Tween 20 1 ml 0.1%
Acknowledgments
We thank K. Furuyama-Tanaka and S. Shimabukuro-Demoto for the technical assistance. This protocol was adapted from Axis-shield’s density gradient protocol S44. This work was supported by the Funding Program for Next Generation World-Leading Researchers (Next Program; for N.T.-S., LS134), grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (for N.T.-S., 21390123, for T.K., 25871165), and a grant from the National Center for Global Health and Medicine (for N.T.-S., 23S001).
References
- Kobayashi, T., Shimabukuro-Demoto, S., Yoshida-Sugitani, R., Furuyama-Tanaka, K., Karyu, H., Sugiura, Y., Shimizu, Y., Hosaka, T., Goto, M., Kato, N., Okamura, T., Suematsu, M., Yokoyama, S. and Toyama-Sorimachi, N. (2014). The histidine transporter SLC15A4 coordinates mTOR-dependent inflammatory responses and pathogenic antibody production. Immunity 41(3): 375-388.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kobayashi, T., Tanaka, T. and Toyama-Sorimachi, N. (2015). Separation of Intracellular Vesicles for Immunoassays. Bio-protocol 5(16): e1571. DOI: 10.21769/BioProtoc.1571.
Category
Immunology > Immune cell isolation > Lymphocyte
Cell Biology > Organelle isolation > Outer membrane vesicles
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








