- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Glucosinolates Determination in Tissues of Horseradish Plant
Published: Vol 5, Iss 16, Aug 20, 2015 DOI: 10.21769/BioProtoc.1562 Views: 8824
Reviewed by: Arsalan DaudiFernanda SalvatoAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Quick Method to Quantify Iron in Arabidopsis Seedlings
Chandan Kumar Gautam [...] Wolfgang Schmidt
Mar 5, 2022 3932 Views

Isolation of Intact Vacuoles from Arabidopsis Root Protoplasts and Elemental Analysis
Chuanfeng Ju [...] Zhenqian Zhang
Mar 5, 2023 2053 Views

High-Performance Liquid Chromatography Quantification of Glyphosate, Aminomethylphosphonic Acid, and Ascorbate in Culture Medium and Microalgal Cells
Juan Manuel Ostera [...] Gabriela Malanga
Apr 5, 2025 1182 Views
Abstract
Glucosinolates (GLS) are secondary metabolites mainly found in plants belonging to the Brassicaceae family, including also horseradish (Armoracia rusticana G. Gaertn., B. Mey. & Scherb), a popular spice with a characteristic pungent flavor due to the abundance of GLS. Such compounds exhibit antibacterial, antifungal, and insecticidal activities, as well as human health properties. Therefore, it is very important to have a full understanding of their levels and profiles in plants. However, the characterization of GLS from horseradish crude extracts is a tough task, due to the complexity of the vegetal matrix and the occurrence of many GLS in trace amounts. Here we describe two alternative effective and rapid methods for GLS characterization in horseradish plants: Liquid chromatography coupled to high resolution mass spectrometry (LC-MS) for determination of intact GLS and HPLC-UV for determination of desulfo-GLS.
Materials and Reagents
- Horseradish tissue (hypogeous and epigeous portion) (see Note 1)
- Methanol (MeOH) (LC/MS grade) (Carlo Erba Reagents, catalog number: 414831 )
- Acetonitrile (ACN) (LC/MS grade) (Carlo Erba Reagents, catalog number: 412342 )
- Ultrapure Milli-Q water
- Liquid nitrogen
- Sinigrin hydrate from horseradish (99%) (Sigma-Aldrich, catalog number: S1647 )
- Rapeseed ERM certified Reference Material containing gluconapin, 4-hydroxyglucobrassicin, glucobrassicanapin, glucobrassicin and gluconaturtiin (Sigma-Aldrich, catalog number: ERMBC367 )
- Glucoiberin (C2 Bioengineering, catalog number: 10-JS 12-05-02 )
- Glucobarbarin (C2 Bioengineering, catalog number: 18-DM 19-10-99 )
- Glucotrapeolin (C2 Bioengineering, catalog number: 16-PM 19-10-99 )
- 70% MeOH (see Recipes)
In addition only for intact GLS determination
- Formic acid (gradient grade) (Sigma-Aldrich, catalog number: F0507 )
- 0.1% Formic acid (HCOOH) (see Recipes)
In addition only for desulfo-method
- DEAE-Sephadex A-25 (formiate form) obtained by using DEAE-Sephadex A-25 (chloride form) (Sigma-Aldrich, catalog number: A25120 ) and imidazole (Sigma-Aldrich, catalog number: 56750 )
- Sulfatase type H-1 (Sigma-Aldrich, catalog number: S-9626 )
- Sulfatase type H-1 (1 to 2.5) (see Recipes)
- DEAE-Sephadex A-25 (formiate form) (see Recipes)
Equipment
- Freeze Dry Systems (e.g. Labconco, model: Freezone 4.5 )
- Laboratory mill
- Disposable 50 ml and 15 ml polypropylene tubes
- 2 ml sample vials
- Water bath: beaker filled with water and placed on a heating device (electric hotplate or similar device).
- Thermometer
- Vortex mixer
- Refrigerated centrifuge (50 ml tubes) (e.g. Heraeus, model: Varifuge F )
- Glass Pasteur pipettes
- 0.22 µm nylon filter (Whatman)
- HPLC system with a photodiode array detector (e.g. Agilent, model: Agilent 1200 HPLC Liquid Chromatrography System)
- HPLC 2 ml glass vials (Phenomenex, model: AR1-3910-12 ) with caps (Phenomenex, model: AR0-8959-13-B )
- Liquid chromatography (LC) coupled with electrospray ionization (ESI) and high resolution mass spectrometry (MS) (e.g. Thermo Fisher Scientific, model: LC-ESI-FTICR MS )
- Intact glucosinolates Discovery C18 column, 250 x 4.6 mm, 5 µm particle size (pore size, 180Å) (Sigma-Aldrich, catalog number: 504971 ), with a Discovery C18 20 x 4 mm security guard cartridge (Sigma-Aldrich, catalog number: 505129 )
- Desulfoglucosinolates (Nucleodur C18 column, 125 mm x 3 mm) (MACHEREY-NAGEL, catalog number: MN760051.30 )
Procedure
Intact GLS
- GLS extraction
- Clean plant material with distilled water and dry with paper towels. Separate roots from epigeous portion, weight and immediately freeze at -80 °C to inhibit myrosinase activity.
- Lyophilize the frozen tissues and grind to a fine powder using a laboratory mill. Chill roots in liquid nitrogen before the lyophilization to allow the crushing.
- Weigh 200 mg of frozen dry material in 50 ml polypropylene tubes and place in a water bath heated with an electric hotplate at 70-80 °C for 10 min. During this process, carry out the following steps: After 1 min add 2 ml of 70% methanol solvent and after 5 min mix with vortex for 20 sec.
- At the end of 10 min remove the polypropylene tube from the water bath and mix again with vortex for 20 sec.
- Centrifuge at 4 °C for 10 min at 2,400 x g.
- Collect the supernatant in a disposable 15 ml polypropylene tube by using glass Pasteur pipettes.
- Extract again the remaining pellet with 2 ml of 10% methanol in water bath heated with an electric hotplate at 70-80 °C for 10 min; after 5 min mix with vortex for 20 sec and the follow the same procedure from step A4-6.
- Combine the supernatants and vortex the extract for 20 sec.
- Filter through 0.22 µm nylon filter and transfer into 2 ml sample vials.
- Clean plant material with distilled water and dry with paper towels. Separate roots from epigeous portion, weight and immediately freeze at -80 °C to inhibit myrosinase activity.
- GLS detection by LC MS
- Inject 25 µl of each sample in LC-ESI-FTICR MS system equipped with Discovery C18 column, 250 x 4.6 mm, 5 µm particle size (pore size, 180 Å), with a Discovery C18 20 x 4 mm security guard cartridge.
Settings:
Flow rate: 1 ml/min
Column temperature: 25 °C
Solvent gradient for chromatographic separation:Time (min) % Solvent A
0.1% HCOOH water% Solvent B
ACN0 90 10 10 76 24 12 40 60 15 90 10 20 90 10
Connect the LC system to the mass spectrometer by a laboratory-made splitter with a split ratio of 1:4 after the analytical column to allow 200 µl/min to enter the ESI source. - Perform full-scan experiments in both the linear trap and the ICR cell. Collect data in profile mode in the range of m/z 50−1,000 by setting:
Ionization mode: negative
Resolution: 100.000 (FWHM) at m/z 400
ESI needle voltage: -4.60 kV
Capillary voltage: -22 V
Temperature of the heated capillary: 350 °C
Sheath gas (N2) flow rate: 80 (arbitrary units)
In these conditions GLS elute with a retention time and molecular exact mass to charge ratio as reported in the following table (Agneta et al., 2012; Agneta et al., 2014):Note: It is possible to use the extracts to quantify GLS by preparing a calibration curve for each GLS using a series of dilution and calculating the area under the peak of each compound.Chemical name Molecular formulae Monoisotopic exact value as [M-H]- (m/z) tR 3-(Methylsulfinyl)propyl-GLS C11H21NO10S3 422.02549 4.3 2-Propenyl-GLS C10H17NO9S2 358.02720 4.4 2-Methylsulfonyl-oxo-ethyl-GLS C10H17NO12S3 437.98402 4.6 3-Butenyl-GLS C11H19NO9S2 372.04285 5.5 1-Methylpropyl-GLS C11H21NO9S2 374.05850 6.2 2-Methylpropyl-GLS C11H21NO9S2 374.05850 6.4 4-Mercaptobuthyl-GLS C11H21NO9S3 406.03057 6.5 7-Methylsulfinylheptyl-GLS C15H29NO10S3 478.08808 7.3 4-Hydroxyindol-3-ylmethyl-GLS C16H20N2O10S2 463.04866 7.4 Unidentified isomer of 4-hydroxyindol-3-ylmethyl-GLS C16H20N2O10S2 463.04866 7.5 2(S)-Hydroxy-2-phenylethyl-GLS
and/or
2(R)-Hydroxy-2-phenylethyl-GLSC15H21NO7S 438.05341 7.8 4-Pentenyl-GLS C12H21NO9SO2 386.05850 7.8 Benzyl-GLS C14H19NO9S2 408.04285 8.5 Indol-3-ylmethyl-GLS C16H20N2O9S2 447.05375 10.3 2-Phenylethyl-GLS C15H21NO9S2 422.05850 12.1 4-Methoxyindol-3-ylmethyl-GLS C17H22N2O10S2 477.06431 12.7 7-Methylthioheptyl-GLS C15H29NO9S3 462.09317 16.7 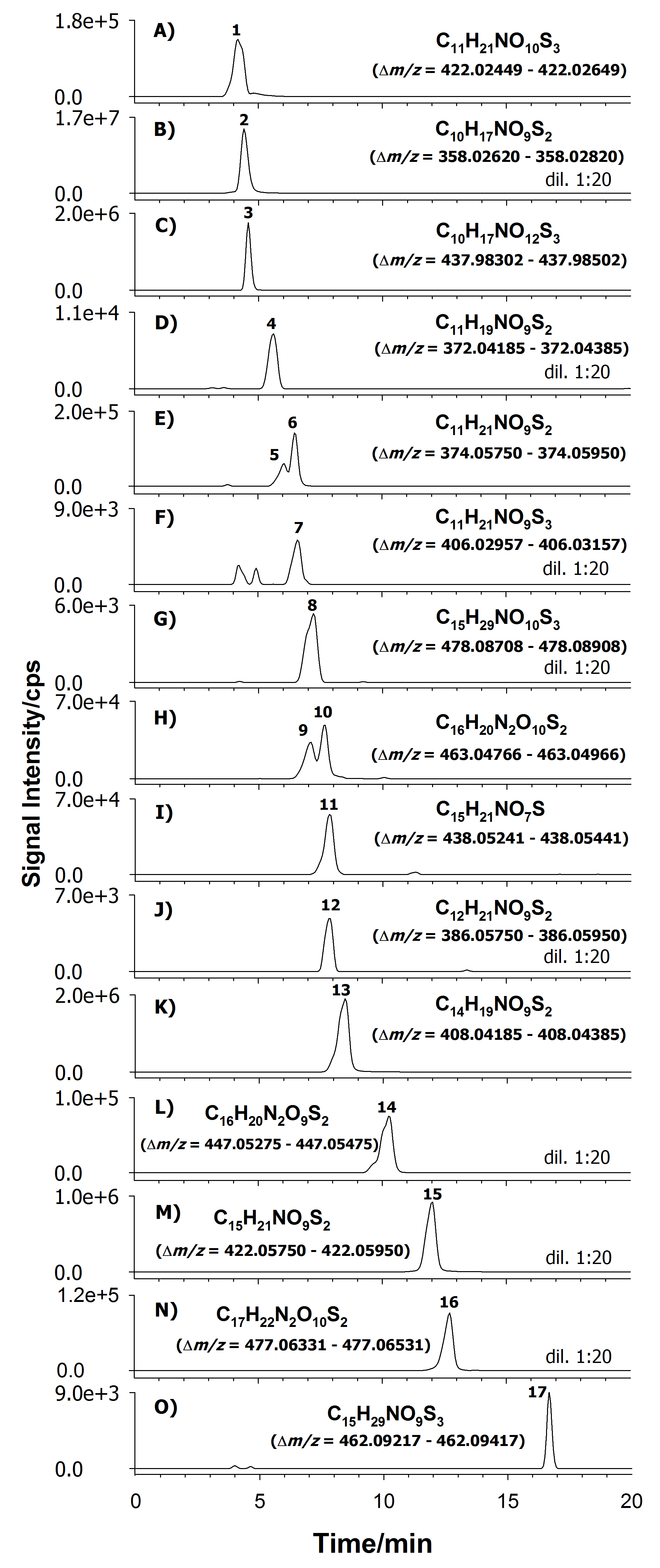
Figure 1. Example of eXtracted Ion Chromatograms (XICs) using high resolution LC-ESI-FTICR MS acquired in negative ion mode of root crude extract of A. rusticana. The ions monitored are displayed in each trace (plots A-O) and correspond to the most abundant deprotonated molecules, [M−H]−, using a restricted window of ±0.0010 m/z unit centered around each selected ion. Peak numbers in bold correspond to: (1) 3-(methylsulfinyl)propyl-GLS; (2) 2-propenyl-GLS; (3)* 2-methylsulfonyl-oxo-ethyl-GLS; (4) 3-butenyl-GLS; (5)* 1-methylpropyl-GLS; (6)* 2-methylpropyl-GLS; (7)* 4-mercaptobuthyl-GLS; (8)* 7-methylsulfinylheptyl-GLS; (9) 4-hydroxyindol-3-ylmethyl-GLS; (10) unidentified isomer of 4-hydroxyglucobrassicin; (11) 2(S)-hydroxy-2-phenylethyl-GLS and/or 2(R)-hydroxy-2-phenylethyl-GLS; (12) 4-pentenyl-GLS; (13) benzyl-GLS; (14) indol-3-ylmethyl-GLS; (15) 2-phenethyl-GLS; (16) 4-methoxyindol-3-ylmethyl-GLS; (17)* 7‐methylthioheptyl-GLS (peaks with asterisk are tentatively assigned). The XIC signals of some peaks were acquired using a sample extract diluted 1:20 with the mobile phase. In bold are indicated also the molecular formula and the range of monoisotopic value as [M-H]– ion (Δ m/z) (Agneta et al., 2014).
- Inject 25 µl of each sample in LC-ESI-FTICR MS system equipped with Discovery C18 column, 250 x 4.6 mm, 5 µm particle size (pore size, 180 Å), with a Discovery C18 20 x 4 mm security guard cartridge.
Desulfo GLS
- GLS extraction
- Follow the procedure of intact GLS step A1-8.
- Transfer 500 µl of the extract on the top of a 20 mg of Sephadex DEAE-A 25 (shortened Pasteur pipet) in the formiate form.
- Wash the column twice with 1 ml of deionized water
- Add 100 µl of sulfatase type H-1 diluted 1 to 2.5 with water to achieve the desulfation and incubate overnight at 39 °C (minimum 16 h) (Möllers et al., 1999)
- Elute desulfatated GLS with 3 x 500 µl deionized water and collect in 15 ml polypropylene tubes.
- Vortex the sample for 20 sec.
- Filter through 0.22 µm nylon filter and transfer into 2 ml sample vials.
- Follow the procedure of intact GLS step A1-8.
- GLS detection by HPLC
- Inject 25 µl of each sample in HPLC system equipped with Nucleodur C18 column, 125 mm x 3 mm.
Settings:
Flow rate: 0.6 ml/min
Column temperature: 35 °C
UV detection: 229
Solvent gradient for chromatographic separation:Time(min) % Solvent A Water % Solvent B ACN 0 99 1 20 80 20 25 80 20 22 99 1
In these conditions GLS elute with a retention time as reported in the following table:aThe response factors values used for the desulfated glucosinolates quantification as reported in EN ISO 9167-1 (1992), with except of following desulfated molecules: 1-methylpropyl-GLS, 2-methylpropyl-GLS and 2(S)-hydroxy-2-phenylethyl-GLS and/or 2(R)-Hydroxy-2-phenylethyl-GLSChemical name tR Relative response factora 3-(Methylsulfinyl)propyl-GLS 2.8 1.07 2-Propenyl-GLS 4.3 1.00 3-Butenyl-GLS 7.3 1.11 1-Methylpropyl-GLS and/or 8.9 1.00b 2-Methylpropyl-GLS 8.9 1.00b 2(S)-Hydroxy-2-phenylethyl-GLS and/or 2(R)-Hydroxy-2-phenylethyl-GLS 10.1 0.98c 4-Pentenyl-GLS 10.9 1.15 Benzyl-GLS 11.8 0.95 Indol-3-ylmethyl-GLS 13.9 0.29 2-Phenylethyl-GLS 16.1 0.95 4-Methoxyindol-3-ylmethyl-GLS 16.5 0.25
bResponse factor of 2-propenyl-GLS as desulfated molecules used for the relative quantification
cExperimental response factor determinate by using the 2(S)-hydroxy-2-phenylethyl-GLS in comparison to 2-propenyl-GLS as desulfated molecules (see Note 2)
- Inject 25 µl of each sample in HPLC system equipped with Nucleodur C18 column, 125 mm x 3 mm.
Notes
- Plant tissues were collected from a field collection of the Institute of Plant Genetics, National Research Council, Thematic Centre for the Preservation of Mediterranean Biodiversity, located in Policoro (MT) (40° 17′ 30″ N, 16° 65′ 16″ E), where many accessions of horseradish, previously collected from various villages of the internal areas of the Basilicata region, are maintained and vegetatively propagated (for details see Sarli et al., 2012).
- It is possible to use the extracts to quantify desulfo GLS by preparing a calibration curve for each GLS by using a series of dilution and calculating the area under the peak of each compound.
It is also possible to quantify desulfo GLS by using the response factors reported in the table above, if you add an internal standard after step A3 (Procedure A), and by applying the following formula:
µmol Areadg x RFdg x n
------- = ------------------------
g Areast x RFst X m
Wherein:
Areadg is the peak area of the desulfoglucosinolate, Areast is the peak area of internal standard, RFdg is the response factor of the corresponding desulfoglucosinolate, RFst is the response factor of the internal standard, n is the quantity, in µmol, of the internal standard added to the sample, m is the mass, in g, of the sample.
Recipes
- 70% MeOH
70 ml of MeOH
30 ml of Milli-Q water - 0.1% Formic acid (HCOOH)
1 ml of Formic acid
999 ml of Milli-Q water - Sulfatase type H-1 (1 to 2.5)
1 ml of sulfatase type H-1
1.5 ml of Milli-Q water - DEAE-Sephadex A-25 (formiate form)
Convert the chloride form to the formiate form by adding for each column 500 µl of 6 mM imidazole formiate (dissolve 40 g imidazole in 100 ml 30% formic acid) and then rinsing the column with water.
References
- Agneta, R., Lelario, F., De Maria, S., Möllers, C., Bufo S. A. and Rivelli A. R. (2014). Glucosinolate profile and distribution among plant tissues and phenological stages of field-grown horseradish. Phytochem 106: 178-187.
- Agneta, R., Rivelli, A. R., Ventrella, E., Lelario, F., Sarli, G. and Bufo, S. A. (2012). Investigation of glucosinolate profile and qualitative aspects in sprouts and roots of horseradish (Armoracia rusticana) using LC-ESI-hybrid linear ion trap with Fourier transform ion cyclotron resonance mass spectrometry and infrared multiphoton dissociation. J Agric Food Chem 60(30): 7474-7482.
- EN ISO 9167-1 (1992). Rapeseed-Determination of glucosinolates content-Part 1: Method using high-performance liquid chromatography.
- Möllers, C., Nehlin, L., Glimelius, K. and Iqbal M. C. M. (1999). Influence of in vitro culture conditions on glucosinolate composition of microspore-derived embryos of Brassica napus. Physiol Plantarum 107(4): 441-446.
- Sarli, G., De Lisi, A., Agneta, R., Grieco, S., Ierardi, G., Montemurro, F., Negro, D., Montesano, V. (2012). Collecting horseradish (Armoracia rusticana, Brassicaceae): local uses and morphological characterization in Basilicata (Southern Italy). Genet Resour Crop Ev 59(5): 889-899.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lelario, F., De Maria, S., Agneta, R., Mӧllers, C., Bufo, S. A. and Rivelli, A. R. (2015). Glucosinolates Determination in Tissues of Horseradish Plant. Bio-protocol 5(16): e1562. DOI: 10.21769/BioProtoc.1562.
Category
Plant Science > Plant biochemistry > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









