- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
High-throughput Quantification of Ammonium Content in Arabidopsis
Published: Vol 5, Iss 16, Aug 20, 2015 DOI: 10.21769/BioProtoc.1559 Views: 11531
Reviewed by: Arsalan DaudiSamik BhattacharyaSaminathan Thangasamy

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Putrescine Biosynthesis Inhibition in Tomato by DFMA and DFMO Treatment
Emma Fernández-Crespo [...] Gemma Camañes
Nov 5, 2016 9042 Views

Evaluation of Root pH Change Through Gel Containing pH-sensitive Indicator Bromocresol Purple
Aparecida L. Silva [...] Daniel S. Moura
Apr 5, 2018 10021 Views

Quantification of the Humidity Effect on HR by Ion Leakage Assay
Musoki Mwimba and Xinnian Dong
Apr 5, 2019 6110 Views
Abstract
This protocol is a simple colorimetric assay for internal ammonium quantification in aqueous extracts from plant tissues. The method is based on the phenol hypochlorite assay (Berthelot reaction):
NH4+ + hypochlorite + OH- + phenol → indophenol
The oxidation of indophenol caused by phenol oxidation is a blue dye that is quantified at 635 nm in a spectrophotometer. Per ammonium molecule one molecule of indophenol is formed. The protocol described here is for Arabidopsis thaliana (A. thaliana) leaves and roots, although it is also valid for other plants species.
Materials and Reagents
- 4-weak old A. thaliana leaves and roots
- Ultrapure water (MilliQ) (EMD Millipore)
- Ice
- Liquid N2
- 4 mm diameter glass beads (Glaswarenfabrik Karl Hecht) (VWR International, catalog number: 201-0278 )
- EVA Capband for capping 8 tubes (Micronic, catalog number: MP227B1 )
- Flat bottom spectrophotometer microplates (Deltalab, catalog number: 900011.1 )
- 1.2 ml 96-well storage plate (Thermo Fisher Scientific, catalog number: AB-0564 )
- Sodium phenolate or sodium phenoxide trihydrate (Sigma-Aldrich, catalog number: 318191 )
- Sodium nitroprusside dihydrate (Sigma-Aldrich, catalog number: s0501 )
- Commercial bleach or Sodium Hypochlorite solution 10% (Panreac Applichem, catalog number: 211921 )
- Solution A (see Recipes)
- Solution B (see Recipes)
- Solution C (see Recipes)
- 10 mM NH4+ stock (for standard curve) (see Recipes)
Equipment
- TissueLyser (Retsch, model: MM400 )
- TissueLyser Adapter Set 2 x 96 (QIAGEN, catalog number: 69984 )
- Plate centrifuge (Sigma, model: 2-16K )
- Drying oven
- Absorbance microplate reader (Biotek PowerWave X 340 Microplate Spectrophometer)
Procedure
- Standard curve
- Prepare dilutions from the 10 mM NH4+ stock in a 0.05 mM - 1 mM range and use ultrapure water as blank. For example: blank; 0.05 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM.
- Material harvest and homogenization
- In a 96 deepwell (1.2 ml) plate, place 2 glass beads (4 mm diameter) into each well.
- Weight approximately 20 mg of fresh leaves or roots and place them in a microplate deepwell half-submerged in liquid nitrogen. Store at -80 °C until use.
- Grind frozen tissue with a TissueLyser two times for 60 sec at 27 Hz frequency. The plates are coupled to the TissueLyser with the TissueLyser Adapter Set 2 x 96. (Ensure sealing of the wells during homogenization).
- In a 96 deepwell (1.2 ml) plate, place 2 glass beads (4 mm diameter) into each well.
- Extraction
- Add 500 μl ultrapure water per well.
- Grind again as in step B3 (make sure the water is not frozen before grinding).
- Incubate samples at 80 °C in a drying oven for 10 min (the incubation can be also done in a water bath).
- Centrifuge the plate at 4,000 rpm and 4 °C for 20 min in a plate centrifuge.
- Recover supernatants (400 μl) in a new 96 deepwell plate and keep the samples on ice.
- Add 500 μl ultrapure water per well.
- Ammonium content measurement
- Place 50 µl of extract (or 50 µl of standards for the calibration curve) per well, in a 96 well spectrophotometer microplate.
- Add 100 µl of Solution A, 50 µl of Solution B and finally 100 µl of Solution C to every sample.
- Make blank using 50 µl of water instead of the plant extract.
- Incubate 30 min at room temperature.
- Read absorbance at 635 nm in a spectrophotometer (Figure 1).
- Making three technical replicates per sample or standard is recommended.
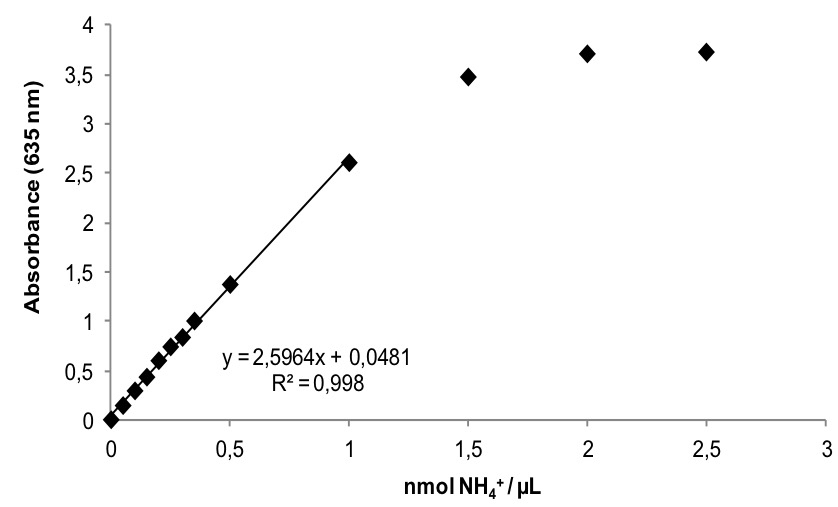
Figure 1. Representative ammonium standard curve. When ammonium content is higher than 1 nmol NH4+ per microliter of reaction mixture absorbance starts to get saturated. Linear regression is performed before arriving to saturation.
- Place 50 µl of extract (or 50 µl of standards for the calibration curve) per well, in a 96 well spectrophotometer microplate.
Data analysis
Subtract the blank absorbance from the absorbance of every sample and calculate the ammonium concentration with the standards calibration curve. Finally, express the ammonium content in a fresh weight basis. These calculations might be also done directly by the microplate reader software assigning to every well a category for example, blank, standard or sample.
Notes
- This protocol can be easily adapted to Eppendorf tubes and spectrophotometer cuvettes instead of 96-well plates just scaling the volumes. Similarly, the homogenisation of the samples might be done with a mortar and pestle instead of the TissueLyser.
- If possible, we advice to do every 96-well plate step using multi-pipettes to save time and thus to gain reproducibility among samples.
Recipes
- Solution A
0.33 M sodium phenolate
Prepare 2 M NaOH dissolving 8 g NaOH in 100 ml of ultrapure water
Mix 2.8 g of sodium phenolate with 4 ml of 2 M NaOH in a final volume of 50 ml with ultrapure water
Ensure pH of the solution is around 13
- Solution B
0.02% sodium nitroprusside
Dissolve 20 mg of sodium nitroprusside in 100 ml of water
Prepare just before using
- Solution C
2% sodium hypochlorite
Dilute 1 ml of commercial bleach in 49 ml of ultrapure water
- 10 mM NH4+ stock (for standard curve)
Dissolve 33.025 mg of (NH4)2SO4 in 50 ml of ultrapure water (of course NH4Cl can also be used as standard)
Acknowledgments
This protocol is adapted from Sarasketa et al. (2014) and based on the methodology reported by Solorzano (1969). This work was supported by the Basque Government (IT526-10), the UPV/EHU (EHUA14/14), the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007–2013) under REA grant agreement number 334019 and MINECO (BIO2014-56271-R).
References
- Sarasketa, A., Gonzalez-Moro, M. B., Gonzalez-Murua, C. and Marino, D. (2014). Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J Exp Bot 65(20): 6023-6033.
- Solorzano L. (1969). Determination of ammonium in natural waters by the phenol-hypochlorite method. Limnol Oceanogr 14: 799-801.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Vega-Mas, I., Sarasketa, A. and Marino, D. (2015). High-throughput Quantification of Ammonium Content in Arabidopsis. Bio-protocol 5(16): e1559. DOI: 10.21769/BioProtoc.1559.
Category
Plant Science > Plant metabolism > Nitrogen
Plant Science > Plant physiology > Ion analysis
Biochemistry > Other compound > Ion > Nitrogen
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









