- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
FACS-based Satellite Cell Isolation From Mouse Hind Limb Muscles
Published: Vol 5, Iss 16, Aug 20, 2015 DOI: 10.21769/BioProtoc.1558 Views: 13522
Reviewed by: Jalaj GuptaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1255 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views
Abstract
Fluorescence Activated Cell Sorting (FACS) is a sensitive and accurate method for purifying satellite cells, or muscle stem cells, from adult mouse skeletal muscle (Liu et al., 2013; Sacco et al., 2008; Tierney et al., 2014). Mechanical and enzymatic digestion of hind limb muscles releases mononuclear muscle cells into suspension. This protocol employs fractionation strategies to deplete cells expressing the cell surface markers CD45, CD31, CD11b and Ly-6A/E-Sca1, both by magnetic separation and FACS-based exclusion, and positively select for cells expressing a7-integrin and CD34. This enables the researcher to successfully enrich satellite cells that uniformly express the paired-box transcription factor Pax7 and are capable of long-term self-renewal, skeletal muscle repair and muscle stem cell pool repopulation.
Materials and Reagents
- Laboratory mice, C57BL/6 strain, 2-4 months of age
Note: All protocols have been approved by the Sanford-Burnham Medical Research Institute Animal Care and Use Committee. - Isoflurane (Santa Cruz Biotechnology, catalog number: sc-363629Rx )
- Ham’s F-10 media (Life Technologies, Gibco®, catalog number: 11550-043 )
- Horse serum (Life Technologies, Gibco®, catalog number: 16050-122 )
- Collagenase type II (Life Technologies, Gibco®, catalog number: 17101-015 )
- Dispase II (Roche Diagnostics, catalog number: 04942078001 )
- Phosphate buffered saline (PBS) (pH 7.4) (Life Technologies, catalog number: 10010-023 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E6758 )
- Goat serum (Life Technologies, Gibco®, catalog number: 16210-072 )
- Antibodies
- Biotin CD45 (clone 30-F11, 0.5 mg/ml) (BD Biosciences, catalog number: 553078 )
- Biotin CD31 (clone 390, 0.5 mg/ml) (eBioscience, catalog number: 13-0311-85 )
- Biotin CD11b (clone M1/70, 0.5 mg/ml) (BD Biosciences, catalog number: 553309 )
- Biotin Ly-6A/E-Sca1 (clone E13-161.7, 0.5 mg/ml) (BD Biosciences, catalog number: 553334 )
- 7-integrin/PE or FITC (clone R2F2, 1 mg/ml) (AbLabs, catalog number: 53-0010-01 )
- CD34/Alexa Fluor 647 (clone RAM34, 0.2 mg/ml) (BD Biosciences, catalog number: 560230 )
- Streptavidin APC-Cy7 (0.2 mg/ml) (BD Biosciences, catalog number: 554063 )
- Streptavidin microbeads (Miltenyi Biotec, catalog number: 130-048-101 )
- FxCycle Violet stain (DAPI, 0.5 mg/ml) (Life Technologies, InvitrogenTM, catalog number: F10347 )
- Biotin CD45 (clone 30-F11, 0.5 mg/ml) (BD Biosciences, catalog number: 553078 )
- Media (see Recipes)
- Stock collagenase type II solution (see Recipes)
- Digestion media I (see Recipes)
- Digestion media II (see Recipes)
- FACS buffer (see Recipes)
Equipment
- Isoflurane vaporizer, supply gas (oxygen), flowmeter and induction chamber
- TC-treated culture dish (10 cm) (Corning Incorporated, catalog number: 430167 )
- Polypropylene centrifuge tubes, sterile (50 ml) (BD Biosciences, catalog number: 352098 )
- Polypropylene centrifuge tubes, sterile (15 ml) (BD Biosciences, catalog number: 352096 )
- LS columns (Miltenyi Biotec, catalog number: 130-042-401 )
- Cell strainers (70 μm) (Thermo Fisher Scientific, catalog number: 22363548 )
- Syringes with 20G x 1’’ needles (BD Biosciences, catalog number: 309644 )
- 5 ml FACS round-bottom tubes (BD Biosciences, Falcon®, catalog number: 352063 )
- Tools for muscle dissection and mincing: Razor blades and/or small scissors, forceps
- Tissue culture laminar flow hood
- Shaking water bath
- Standard temperature-controlled table-top centrifuge
- MACS magnetic separator and multi-stand (Miltenyi Biotec, catalog number: 130-042-302 and 130-042-303 )
- Flow cytometer [, (FACSAria cell sorter equipped with 488, 405 and 633 nm lasers) (BD Biosciences)
Note: Color combinations can be adjusted to match the laser combinations available.
Software
- FlowJo software (Tree Star, optional)
Procedure
- Warm media in 37 °C water bath.
- On a surgical bench, harvest the hind limb muscles; place all muscles from one hind limb in a 10 cm dish with 4 ml pre-warmed media (one mouse at a time, keep isolated muscles at 37 °C) (Video 1).
- Anesthetize mouse by isoflurane inhalation and sacrifice by cervical dislocation.
- Remove skin covering hind limb muscles.
- Sever the Achilles tendon and separate the gastrocnemius and soleus.
- Sever the distal tendons of the anterior compartment muscles including the tibialis anterior and extensor digitorum longus, use a razor blade to separate from the tibia.
- Sever the quadriceps tendon, use razor blade to separate the quadriceps from the femur.
Video 1. Hind limb muscle isolation - Anesthetize mouse by isoflurane inhalation and sacrifice by cervical dislocation.
- Mince muscles from one hind limb in the 10 cm dish with media using 2 razor blades into small pieces (1-2 plates at a time, keep all other samples at 37 °C).
- Once all muscles are minced, use forceps to transfer muscle pieces from each dish into a 15 ml centrifuge tube. Pieces should stick together and be easily picked up as one or two aggregates and placed into the tube (Figures 1 and 2).
- Using a 5 ml pipet, collect the media from the dish and any muscle pieces that were not picked up with forceps and add it to the 15 ml centrifuge tube.
- Wash the plate with 5 ml media and add it to the 15 ml centrifuge tube. Each tube now contains 9 ml media and the muscle pieces.
- Add 1 ml stock collagenase solution to reach a final volume of 10 ml of digestion media I.
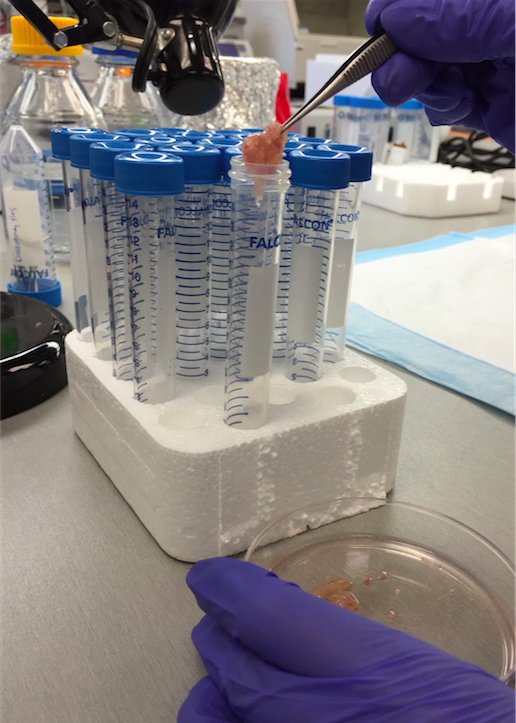
Figure 1. Minced muscle transfer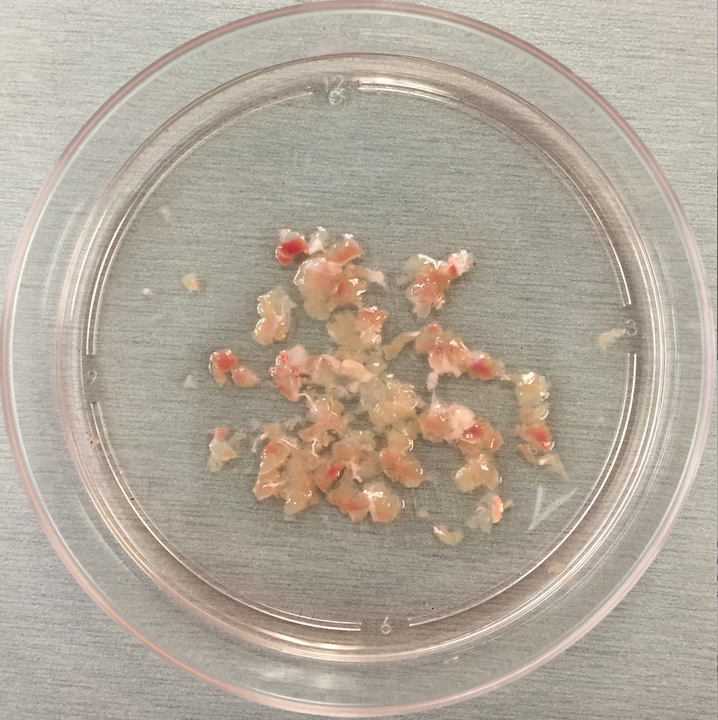
Figure 2. Mincing - Using a 5 ml pipet, collect the media from the dish and any muscle pieces that were not picked up with forceps and add it to the 15 ml centrifuge tube.
- Place samples on a shaking water bath, warmed to 37 °C, at 175-325 rpm for 90 min. Alternatively, if a shaking water bath is not available, place samples in 37 °C incubator and mix contents manually by inverting the tube every 15-20 min. However, this is likely to reduce yield due to sub-optimal digestion.
- Centrifuge samples at 300 x g for 5 min at RT.
- Aspirate supernatant and resuspend in 10 ml digestion media II.
- Briefly vortex samples and return to the 37 °C shaking water bath at 200 rpm for 30 min.
- Pipet up/down to break up muscle pieces and pass each sample through a 10 ml syringe with 20 G needle 10 times to further release all mononuclear cells into suspension. No visible muscle pieces should remain once completed.
- Transfer samples to a 50 ml centrifuge tube, washing each 15 ml centrifuge tube 2 times with 12 ml media.
- Centrifuge samples at 300 x g for 5 min at 4 °C.
- Aspirate supernatant, resuspend samples in 10 ml media and pass through a 70 μm cell strainer atop a fresh 50ml centrifuge tube; wash the original tube 2 times with 12 ml media.
- Centrifuge samples at 300 x g for 5 min at 4 °C.
- Aspirate supernatant, resuspend samples in 600 μl media and transfer to 15 ml centrifuge tube, then wash the original tube with 400 μl media. Samples from both legs can be combined into one tube at this step such that the sort sample contains cells from all muscles of one mouse in 2 ml media.
- Add 10 μl cell suspension from the 2 ml sort sample in 500 μl media and the following volumes of antibody to separate 5 ml FACS tubes for single color controls:
- No antibody (unstained control)
- 1 μl FxCycle Violet stain (DAPI)
- 1 μl α7-integrin/PE or FITC
- 4 μl CD34/Alexa Fluor 647
- 1 μl biotin CD45
- No antibody (unstained control)
- Add the following volumes of primary antibodies to together the original 2 ml sort samples:
- 5 μl biotin CD45
- 10 μl biotin CD11b
- 10 μl biotin CD31
- 10 μl biotin Ly-6A/E-Sca1
- 5 μl biotin CD45
- Gently vortex samples and incubate on ice for 20 min.
- Add 10 ml chilled media to sort samples or 3 ml FACS buffer to single color controls.
- Centrifuge samples at 300 x g for 5 min at 4 °C.
- Aspirate supernatant, resuspend sort samples in 1.5 ml media or single color controls in 500 μl FACS buffer.
- Add 1 μl streptavidin APC-Cy7 to the biotin CD45 single color control and incubate on ice for 20 min.
- Add the following secondary antibodies to sort samples (microbeads first, then remaining antibodies):
- 150 μl streptavidin microbeads
- 10 μl α7-integrin/PE or FITC
- 30 μl CD34/Alexa Fluor 647
- 10 μl streptavidin APC-Cy7
- 150 μl streptavidin microbeads
- Gently vortex samples and incubate on ice for 20 min.
- Add 10 ml chilled media to sort samples or 3 ml FACS buffer to the biotin CD45 single color control.
- Centrifuge samples at 300 x g for 5 min at 4 °C.
- Pre-load LS columns (one for each sample) for depletion with 3 ml media.
- Aspirate supernatant, resuspend sort samples in 1 ml media or the biotin CD45 single color control in 500 μl FACS buffer.
- Load each sort sample onto an LS column placed on a MACS magnetic separator, washing the original centrifuge tube 2 times with 3-4 ml media (keeping samples on ice as much as possible).
- Centrifuge samples at 300 x g for 5 min at 4 °C.
- Aspirate supernatant, resuspend sort samples in 2 ml FACS buffer transferred to 5 ml FACS tubes and add 2 μl FxCycle Violet stain (DAPI) to each sample.
- Keep on ice until ready for sorting on a BD Biosciences FACSAria cell sorter (Figure 3).
a.Cell should be sorted at 20 psi through a 100 μm nozzle.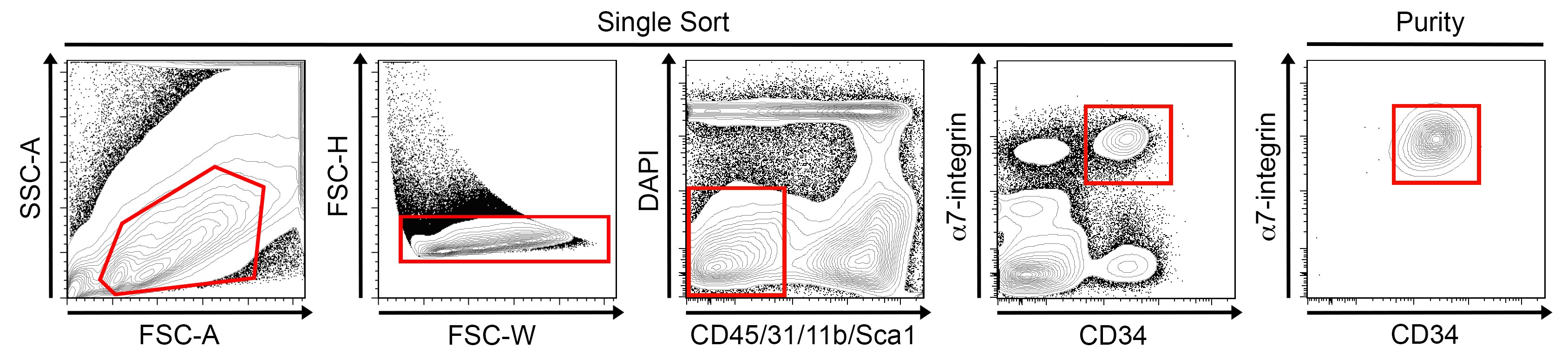
Figure 3. FACS-based satellite cell isolation. Representative FACS plots and gating schemes to purify mononucleated satellite cells both negative for CD45, CD31, CD11b and Sca1 and expressing a7-integrin and CD34 (left). Representative FACS plot showing a7-integrin and CD34 expression in re-sorted satellite cells.
Notes
- During the muscle dissection and mincing steps, look for and discard large pieces of thick white tendons that do not efficiently get digested and may clog the syringe and needle at step 9.
- Razor-based mincing prior to enzymatic digestion is a critical step in the protocol; mincing into large pieces will result in an incomplete digestion of the tissue while mincing into small pieces will lower yield.
- Supernatant from step 7 can be collected, diluted further with cold PBS, centrifuged at 400 x g for 8 min and kept on ice until added back to main sample prior to filtering at step 12 to potentially increase yield.
- Cell suspensions following tissue digestion should be kept on ice or at 4 °C to slow satellite cell activation and sorted as quickly as possible following sample preparation, particularly if analysis is planned for freshly isolated cells when presumed quiescent.
- FxCycle Violet Stain (DAPI) can be used at 300-900 μg/ml before increased autofluorescence is observed. Alternatively, propidium iodide can be used to discriminate between live and dead cells but fluorescence compensation is more difficult with this color scheme.
- Cellular yield from one adult mouse, C57BL/6 strain, 2-4 months of age should be approximately 150,000-200,000 events by FACS analysis and 75,000-100,000 satellite cells when manually counted.
Recipes
- Media
Ham’s F-10 media and 10% horse serum
0.2 μm sterile filtered and stored at 4 °C - Stock collagenase type II solution
Weigh out and dissolve collagenase powder in media (Recipe 1) for a stock concentration of 7,000 units/ml
Units per milligram vary by lot and calculations need to be adjusted accordingly
Stock collagenase solution can be aliquoted and stored at -20 °C for up to 6 months or 4 °C for 1 day
Do not freeze/thaw more than one time - Digestion media I
9 ml media (Recipe 1) and 1 ml stock collagenase type II solution (Recipe 2) - Digestion media II
9.86 ml media (Recipe 1)
143 μl stock collagenase type II solution (Recipe 2, 100 units/ml final concentration)
20 units dispase II (powdered form, weighed out immediately prior to use) - FACS buffer
486.5 ml 1x PBS
12.5 ml goat serum (2.5% final) and 1 ml 0.5 M EDTA (1 mM final)
0.2 μm sterile filtered and stored at 4 °C
Acknowledgments
This protocol was adapted and modified from previous work published by both Tom Rando and Helen Blau’s research groups. This work was supported by the Muscular Dystrophy Association MDA Grant 200845, Ellison Medical Foundation New Scholar Award AG-NS-0843-11 and US National Institutes of Health (NIH) grants R01AR064873, R03 AR063328, P30 AR061303 to AS, and US National Institutes of Health (NIH) grant F31 AR065923-01 to MT.
References
- Liu, L., Cheung, T. H., Charville, G. W., Hurgo, B. M., Leavitt, T., Shih, J., Brunet, A. and Rando, T. A. (2013). Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 4(1): 189-204.
- Sacco, A., Doyonnas, R., Kraft, P., Vitorovic, S. and Blau, H. M. (2008). Self-renewal and expansion of single transplanted muscle stem cells. Nature 456(7221): 502-506.
- Tierney, M. T., Aydogdu, T., Sala, D., Malecova, B., Gatto, S., Puri, P. L., Latella, L. and Sacco, A. (2014). STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med 20(10): 1182-1186.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gromova, A., Tierney, M. T. and Sacco, A. (2015). FACS-based Satellite Cell Isolation From Mouse Hind Limb Muscles. Bio-protocol 5(16): e1558. DOI: 10.21769/BioProtoc.1558.
Category
Stem Cell > Adult stem cell > Muscle stem cell
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










