- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of HBV C Protein Phosphorylation in the Cell
Published: Vol 5, Iss 15, Aug 5, 2015 DOI: 10.21769/BioProtoc.1551 Views: 8588
Reviewed by: Smita NairVarpu MarjomakiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorometric Estimation of Viral Thermal Stability
Vamseedhar Rayaprolu [...] Brian Bothner
Aug 5, 2014 13211 Views

Host-regulated Hepatitis B Virus Capsid Assembly in a Mammalian Cell-free System
Kuancheng Liu and Jianming Hu
Apr 20, 2018 7106 Views

Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC)
Dina A. Abdulrahman and Michael Veit
Apr 5, 2025 1856 Views
Abstract
Among the seven serines and one threonine in the carboxyl-terminus of HBV C protein, all but one (serine 183) appear in the context of RxxS/T consensus phosphoacceptor motifs and also overlap with other consensus motifs, such as S/TP, RS, SPRRR, RRRS/T, or RRxS/T, suggesting that various cellular kinases phosphorylate these residues. To determine whether threonine and/or serine (serines 157, 164, 170, 172, 178, and 180, and threonine 162, adw subtype) of HBV C protein are indeed phosphoacceptor residues in cells, Huh7 were transfected with a series of C-protein-expressing mutants, labeled with 32P-orthophosphate for 14 h, and then lysed. The 32Pi-labeled lysates were immunoprecipitated with anti-HBc antibody, and the 32Pi-labeled immunoprecipitated C proteins were detected by autoradiography.
Materials and Reagents
- Huh7 hepatoma cells (Japanese Collection of Research Bioresources Cell Bank, catalog number: JCRB0403 )
- Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies, Gibco®, catalog number: 12800-017 )
- Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 16000-044 )
- Penicillin/streptomycin (Life Technologies, Gibco®, catalog number: 15140-122 )
- Plasmid; HBV WT C protein (STSSSS) in plasmid HBV-P-def of pcDNA3 backbone, phosphoacceptor-site mutants (ATAAAA, AAAAAA, SSSSSS, and ASAAAA) in plasmid HBV-P-def in pcDNA3 backbone, pcDNA3-GFP
- Polyethylenimine (Polysciences, catalog number: 23966 )
- Opti-MEM (Life Technologies, Gibco®, catalog number: 31985-062 )
- OptiMEM (Life Technologies, Gibco®, catalog number: 31985-070 )
- Dialyzed FBS (Life Technologies, Gibco®, catalog number: 26400044 )
- 1 mCi orthophosphate [32Pi] (PerkinElmer Inc., catalog number: NEX053 )
- Polyclonal rabbit anti-HBc antibody (home-made, Jung et al., 2012)
- Protein A/G Plus agarose beads (Calbiochem®, catalog number: IP05 )
- Tris-HCl (pH 8.5) (Sigma-Aldrich, catalog number: T6066 )
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- Nonidet P-40 (Sigma-Aldrich, catalog number: CA630 )
- NaF (Sigma-Aldrich, catalog number: 201154 )
- β-glycerophosphate (USB, catalog number: 155-56-2 )
- Sodium orthovanadate (Sigma-Aldrich, catalog number: s6508 )
- Protease inhibitors (Calbiochem®, catalog number: 535142 )
- Tris-HCl (pH 6.8) (Sigma-Aldrich, catalog number: T6066)
- SDS (Sigma-Aldrich, catalog number: L-3771 )
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Bromophenol blue (Sigma-Aldrich, catalog number: 114391 )
- Glycerol (USB, catalog number: 16374 )
- Lysis buffer (see Recipes)
- 2x sample buffer (see Recipes)
Equipment
- 10-cm dishes (Corning Incorporated, catalog number: 430167 )
- PVDF membranes (Bio-Rad Laboratories, AbD Serotec®, catalog number: 162-0177 )
Procedure
- Huh7 hepatoma cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin under a humidified atmosphere at 37 °C in 5% CO2.
- Cells were passaged every three days (passage at 80~90% confluency) and 2 x 106 Huh7 cells were seeded in 10-cm dishes one day before the transfection.
- Next day, Huh7 cells were transfected using polyethylenimine (PEI).
PEI transfection method:- In a sterile tube, total 10 μg of plasmid DNA was mixed with 500 μl Opti-MEM.
- Add 30 μl of PEI solution (1 μg/1 μl) to DNA-Opti-MEM solution and then vortex immediately.
- Incubate 15 min at room temperature.
- Then add PEI/DNA-Opti-MEM mixture to cells.
- In a sterile tube, total 10 μg of plasmid DNA was mixed with 500 μl Opti-MEM.
- Transfection experiments were repeated at least three times.
- Forty-eight hours after transfection, Huh7 cells were incubated at 37 °C in 5% CO2 incubator for 6 h in 10 ml DMEM containing 10% dialyzed FBS, then labeled at 37 °C in 5% CO2 for additional 14 h with 250 µCi orthophosphate [32Pi] per 107 cells, rinsed with ice-cold phosphate-buffered saline, and lysed with 1 ml of lysis buffer. (Isotope was added directly onto the cells containing DMEM-dFBS.)
- Lysates were transferred to 1.5 ml tube, incubated on ice for 10 min for further lysis, spin down at 13,000 rpm at 4 °C for 2 min, and supernatant was transferred to fresh 1.5 ml tube.
- The supernatant lysates were incubated for 2 h at 4 °C with 1 μl of polyclonal rabbit anti-HBc antibody on the rotator.
- Next, 15 μl of protein A/G Plus agarose beads was added, and the mixture was incubated at 4 °C for 1 h and centrifuged at 1,500 rpm for 10 sec. (Protein A/G Plus agarose bead preparation: Take 15 μl of bead/sample to fresh 1.5 ml tube, add 1 ml lysis buffer, vortex briefly, spin down at 1,500 rpm for 10 sec, and discard supernatant. Repeat above washing procedures 2 more times.)
- The pellets in 1.5 ml tube (containing beads bound to immune complexes of radiolabeled WT or mutant C proteins and anti-HBc antibodies) were washed twice with 1 ml lysis buffer, eluted by boiling (100 °C for 5 min) in 15 μl of 2x sample buffer, and separated by SDS-PAGE on 13.5% gels. Running time was 3 h at 80 voltage.
- Separated proteins were transferred to PVDF membranes (250 mA 1 h 30 min) and subjected to autoradiography.
Representative data
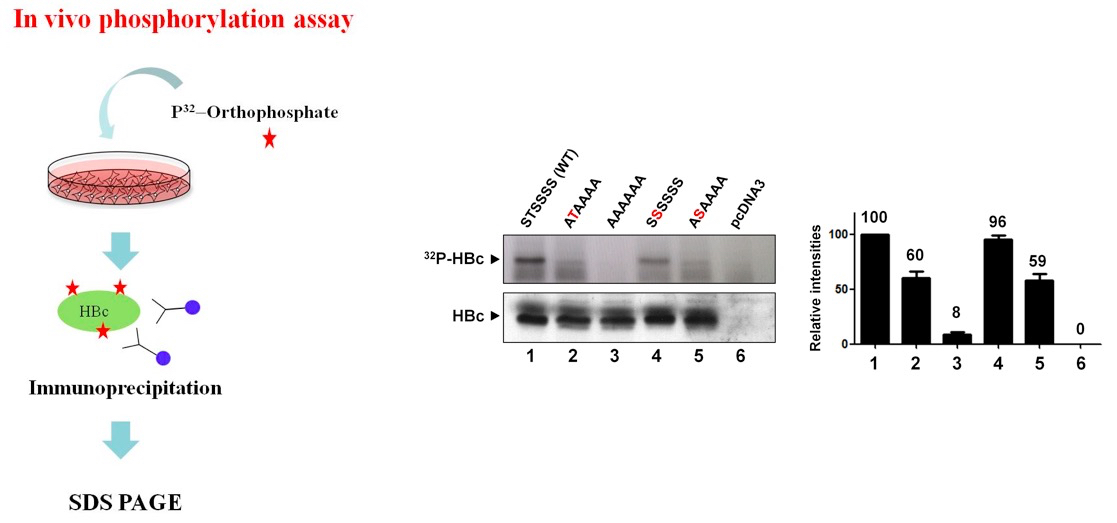
Compared to AAAAAA mutant of which the phosphoacceptor sites were all abolished to alanine, STSSSS (WT) and other mutants were all 32Pi-labeled.
Recipes
- Lysis buffer
50 mM Tris-HCl (pH 8.5)
2 mM EDTA
0.5% Nonidet P-40
50 mM NaF
25 mM β-glycerophosphate
2 mM sodium orthovanadate, and protease inhibitors - 2x sample buffer
100 mM Tris-HCl (pH 6.8)
4% SDS
200 mM β-mercaptoethanol
0.2% bromophenol blue
20% glycerol
Acknowledgments
This work was supported by National Research Foundation Grants funded by the Korean Government (NRF-2012-R1A2A2A01015370).
References
- Jung, J., Kim, H. Y., Kim, T., Shin, B. H., Park, G. S., Park, S., Chwae, Y. J., Shin, H. J. and Kim, K. (2012). C-terminal substitution of HBV core proteins with those from DHBV reveals that arginine-rich 167RRRSQSPRR175 domain is critical for HBV replication. PLoS One 7(7): e41087.
- Jung, J., Hwang, S. G., Chwae, Y. J., Park, S., Shin, H. J. and Kim, K. (2014). Phosphoacceptors threonine 162 and serines 170 and 178 within the carboxyl-terminal RRRS/T motif of the hepatitis B virus core protein make multiple contributions to hepatitis B virus replication. J Virol 88(16): 8754-8767.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jung, J. and Kim, K. (2015). Detection of HBV C Protein Phosphorylation in the Cell. Bio-protocol 5(15): e1551. DOI: 10.21769/BioProtoc.1551.
Category
Microbiology > Microbial biochemistry > Protein > Modification
Biochemistry > Protein > Immunodetection > Western blot
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










