- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of ex vivo Neutrophil Extracellular Traps
Published: Vol 5, Iss 15, Aug 5, 2015 DOI: 10.21769/BioProtoc.1549 Views: 12301
Reviewed by: Ivan ZanoniAchille BroggiMarco Di Gioia

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A New Methodology for the Quantification of Neutrophil Extracellular Traps in Patient Plasma
Bharati Matta [...] Betsy J. Barnes
Jun 20, 2023 3132 Views

Measuring Myeloperoxidase Activity as a Marker of Inflammation in Gut Tissue Samples of Mice and Rat
Nikita Hanning [...] Benedicte Y. De Winter
Jul 5, 2023 2761 Views

Utilizing EdU to Track Leukocyte Recruitment to the Brain
Zoie K. Lipfert [...] David P. Sullivan
Dec 5, 2025 1564 Views
Abstract
Neutrophil extracellular traps (NETs) are fibrous mesh-like, web-like, or string-like structures which are composed of DNA, histones, and granule proteins such as neutrophil elastase or myeloperoxidase. When activated by phorbol myristate acetate, interleukin-8, lipopolysaccharide (LPS), and various pathogens, neutrophils release NETs. We reported that NETs were classified as two distinct forms; cell-free NETs that were released away from neutrophils and anchored NETs that were anchored to neutrophils. In general, extracellular DNAs are used as a surrogate marker of NETs. Here, we describe a protocol regarding quantitative procedures of extracellular DNAs released from ex vivo neutrophils activated by LPS using fluorometric double-stranded DNA (dsDNA) quantification assay.
Materials and Reagents
- Whole blood from wild-type C57/BL6 mice (Japan SLC)
- Whole blood from human volunteers
- LPS (Escherichia coli, serotype 0111:B4) (Sigma-Aldrich, catalog number: L4391 )
- PolymorphprepTM (Axis Shield PoC AS, catalog number: 1114683 )
- RPMI 1640 medium (no phenol red) (Life Technologies, catalog number: 32404-014 )
- Fetal bovine serum (Life Technologies, catalog number: 12483-020 )
- ACK (Ammonium-chloride-potassium) lysing buffer (Lonza, catalog number: 10-548E )
- Quant-iTTM PicoGreen dsDNA Assay Kit (Life Technologies, InvitrogenTM, catalog number: P11496 )
Equipment
- Glass Pasteur pipets (Iwaki brand, Asahi Techno Glass Corporation)
- 96-well plates (TPP Techno Plastic Products AG)
- CO2 Incubator (SANYO)
- Fluorescence microplate reader (Perkin Elmer, model: 2030ARVOX )
Procedure
- Isolation of human neutrophils
- Venous blood (6 ml each) was obtained from healthy human volunteers.
- Neutrophils were isolated by density gradient centrifugation using PolymorphprepTM according to the manufacturer's instructions.
- EDTA anti-coagulated blood (the optimal concentration is 1.5 mg per ml of blood) was layered onto 6 ml Polymorphprep solution and centrifuged at 500 x g for 30 min.
- The granulocyte fraction was carefully harvested using a glass Pasteur pipette.
- Wash the pellet containing granulocytes with 2 ml phosphate buffered saline (PBS).
- Centrifuge at 400 x g for 10 min, and resuspend the pellet in 1 ml PBS.
- When present, erythrocytes were lysed with ACK lysing buffer.
- 1 ml ACK lysing buffer was added to the pellet with residual erythrocytes.
- Incubate at room temperature for 5 min with occasional pipetting.
- Wash the pellet containing granulocytes with 2 ml PBS.
- Centrifuge at 400 x g for 10 min.
- Neutrophils were resuspended in 1 ml of RPMI 1640 without phenol red supplemented with 1% fetal bovine serum.
- Final neutrophil concentration was determined by hemacytometer. Approximately 0.5~1.0 x 107/ml of neutrophils will be obtained.
- Neutrophil purity was confirmed to be routinely >90%, as assessed by May-Grünwald Giemsa staining on the blood smear.
- In brief, immerse the air-dried smear slide in 1 ml May-Grunwald solution for 1 min.
- Add an equal part of phosphate buffer and incubate for 3 min.
- Pour off the stain and wash the slide with tap water.
- Immerse in the 8% Giemsa solution for 20 min.
- Wash the slide with tap water and air-dry.
- Venous blood (6 ml each) was obtained from healthy human volunteers.
- Isolation of murine leukocytes
- Heparinized blood was withdrawn from the inferior vena cava of anesthetized wild-type C57/BL6 mice.
- In brief, open the abdomen and identify the inferior vena cava between the kidneys.
- Use a 25 gauge needle and a 1 ml syringe filled with 50 µl heparin for the prevention of blood coagulation.
- Insert the needle into the vein and draw blood slowly until the vein collapses.
- Approximately 500 µl blood will be obtained.
- ACK lysing buffer was used to lyse erythrocytes.
- 5 ml ACK lysing buffer was added to 500 µl of murine whole blood.
- Incubate at room temperature for 5 min with occasional gentle shaking.
- Centrifuge at 400 x g for 10 min.
- Discard the supernatant containing lysed erythrocytes carefully.
- (If necessary, repeat steps B6-10.)
- Wash the pellet with 2 ml PBS.
- Centrifuge at 400 x g for 10 min, and resuspend the pellet in the 1ml of the above mentioned medium (step A12).
- After ACK treatment, the blood cells that remained included white blood cells (leukocytes) and platelets.
- Final leukocyte concentration was determined by hemacytometer. Approximately 1.0~2.0 x 106/ml of neutrophils will be obtained.
- Heparinized blood was withdrawn from the inferior vena cava of anesthetized wild-type C57/BL6 mice.
- Neutrophil activation by LPS
- Human neutrophils or murine leukocytes obtained from wild-type C57/BL6 mice were suspended in the above-mentioned medium (step A12).
- They were seeded to the 96-well plate (first plate) at a density of 1 x 104 cells per well (100 µl).
- They were stimulated with LPS at indicated concentrations (2, 20, 100, and 200 µg/ml). The plates were placed in a humidified incubator at 37 °C with CO2 (5%) for 6 h.
- Human neutrophils or murine leukocytes obtained from wild-type C57/BL6 mice were suspended in the above-mentioned medium (step A12).
- Quantification of cell-free NETs
- Since cell-free NETs were released away from neutrophils, they were quantified as extracellular DNAs in culture supernatants.
- The culture supernatant of each well (100 µl) was collected and directly transferred to another 96-well plate (second plate). A centrifugation is not needed.
- Cell-free NETs in culture supernatants were quantified as dsDNA in the culture supernatants using a Quant-iTTM PicoGreen dsDNA Assay Kit.
- In brief, an equal volume of PicoGreen reagent (100 µl) was added to each well containing culture supernatants (100 µl).
- Plates were incubated for 5 min at room temperature with protecting from light.
- The PicoGreen dye that was bound to dsDNA was measured using a fluorescence microplate reader. The following settings were used to read the PicoGreen results: Excitation wavelength and bandwidth were 485 nm and 14 nm, respectively. Emission wavelength and bandwidth were 535 nm and 25 nm, respectively.
- The DNA concentration was calculated using a standard curve generated from a series of Lambda DNA standard (100 µg/ml) provided by the manufacturer.
- Since cell-free NETs were released away from neutrophils, they were quantified as extracellular DNAs in culture supernatants.
- Quantification of anchored NETs
- To quantify anchored NETs that were anchored to neutrophils, the first plate was used after removal of culture supernatants.
- Fresh medium (100 µl) was added to each well of the first plate. An equal volume of PicoGreen reagent (100 µl) was added to each well to quantify anchored NETs.
- Plates were incubated for 5 min at room temperature protected from light.
- The PicoGreen dye that was bound to dsDNA was measured using a fluorescence microplate reader.
- The DNA concentration was calculated using a standard curve generated from a series of Lambda DNA standard (100 µg/ml) provided by the manufacturer.
- To quantify anchored NETs that were anchored to neutrophils, the first plate was used after removal of culture supernatants.
Representative data
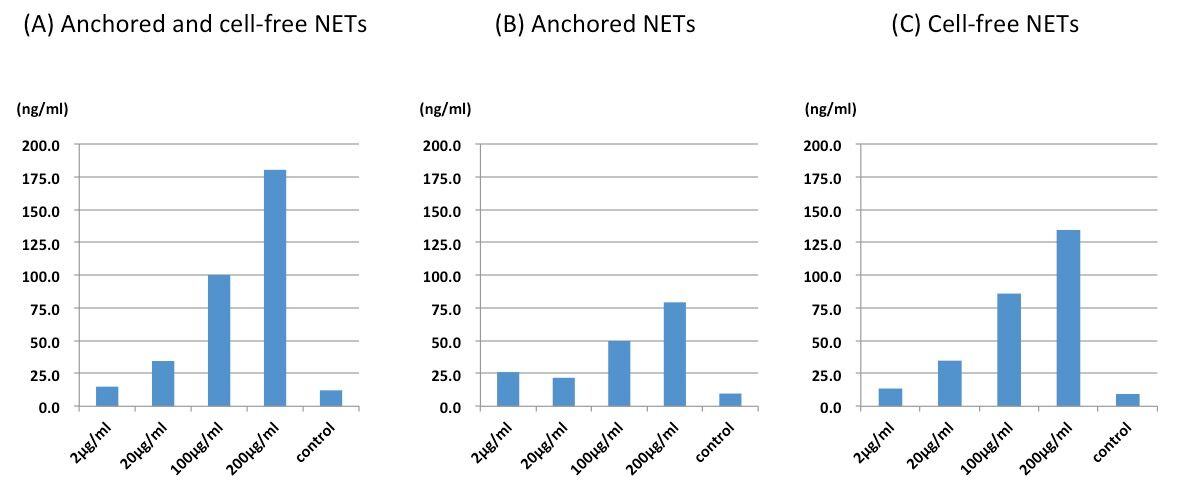
Figure 1. The representative data of ex vivo NETs released from human neutrophils activated by LPS. (A). The DNA concentration at the indicated dose of LPS was shown as the sum of the amount of anchored and cell-free NETs. (B). The DNA concentration of anchored NETs was shown at the indicated dose of LPS on the first plate. Anchored NETs were anchored to neutrophils adhering to the plate. (C). The DNA concentration of cell-free NETs was shown at the indicated dose of LPS. They were transferred to the second plate as the supernatant of the first plate.
Notes
Gentle aspiration of the supernatant should be needed to avoid the contamination of anchored NETs into the second plate.
Acknowledgments
This protocol was adapted from the previously reported in Tanaka et al. (2014). This work was partly supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (KAKENHI 25462052 to K.T.).
References
- Tanaka, K., Koike, Y., Shimura, T., Okigami, M., Ide, S., Toiyama, Y., Okugawa, Y., Inoue, Y., Araki, T., Uchida, K., Mohri, Y., Mizoguchi, A. and Kusunoki, M. (2014). In vivo characterization of neutrophil extracellular traps in various organs of a murine sepsis model. PLoS One 9(11): e111888.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tanaka, K., Okigami, M., Toiyama, Y., Okugawa, Y., Inoue, Y., Araki, T., Mohri, Y., Mizoguchi, A. and Kusunoki, M. (2015). Quantification of ex vivo Neutrophil Extracellular Traps. Bio-protocol 5(15): e1549. DOI: 10.21769/BioProtoc.1549.
Category
Immunology > Immune cell function > Neutrophil
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









