- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mouse Retinal Whole Mounts and Quantification of Vasculature Protocol
Published: Vol 5, Iss 15, Aug 5, 2015 DOI: 10.21769/BioProtoc.1546 Views: 17343
Reviewed by: Shai BerlinSoyun KimAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Human Endothelial Cell Spheroid-based Sprouting Angiogenesis Assay in Collagen
Fabian Tetzlaff and Andreas Fischer
Sep 5, 2018 21480 Views

Isolation of Stem Cells, Endothelial Cells and Pericytes from Human Infantile Hemangioma
Lan Huang and Joyce Bischoff
Jan 20, 2020 4962 Views

An Improved Protocol for the Matrigel Duplex Assay: A Method to Measure Retinal Angiogenesis
Kathleen C. Brown [...] Piyali Dasgupta
Dec 5, 2023 1915 Views
Abstract
Angiogenesis is the formation of new blood vessels from a pre-existing vascular bed. It is a multi-step process beginning with enzymatic degradation of the capillary basement membrane, followed by endothelial cell (EC) proliferation, migration, tube formation, assembly of a new basement membrane, and pericyte stabilization. Aberrant angiogenesis plays a major role in the pathogenesis of many diseases. The regulation of this complex process is an important therapeutic target. Success in this pursuit, however, requires the development of in vivo angiogenesis models that provide a reliable and facile platform for mechanistic studies of angiogenic regulation as well as drug development and testing (Carmeliet and Jain, 2011).
Postnatal development of mouse retinal vasculature offers a unique and powerful in vivo angiogenesis model because, unlike other species, mice undergo extensive angiogenesis-dependent maturation of their retinal vessels after birth. As such, this model is also very useful for the mechanistic study of embryonic vascularization (Stahl et al., 2010; Adini et al., 2003).
This protocol describes the steps involved in the whole mount processing of mouse eyes for visualization of the retinal vasculature.
Materials and Reagents
- 10% Formalin solution (Sigma-Aldrich, catalog number: HT5011284L ) or Paraformaldehyde (PFA) (Electron Microscopy Sciences, catalog number: 15710 )
- Triton X-100 (Sigma-Aldrich, catalog number: 93443 )
- Sodium azide (Sigma-Aldrich, catalog number: S2002 )
- Goat serum (Sigma-Aldrich, catalog number: G9023 )
- Phosphate buffer saline (PBS) (Sigma-Aldrich, catalog number: P3813 )
- Antibody: Bandeiraea simplicifolia BS-1 (LEC)-TRIC (Sigma-Aldrich, catalog number: L-5264 ) diluted 1:200
- Mounting medium: ProLong Gold antifade reagents (Life Technologies, catalog number: P36934 )
- Blocking buffer (see Recipes)
Equipment
- Razor Blade
- Shaker-2 min maximum velocity
- Epifluorescence microscope fitted with camera
- Student Dumont #5 Forceps (Fine Science Tools, catalog number: 9115020 )

- Curve Dumont #7 Fine Forceps (Fine Science Tools, catalog number: 1127420 )

- Micro dissecting scissors (Fine Science Tools, catalog number: 1501810 )

- Microscope cover glasses (Thermo Fisher Scientific, Menzel-Gläser, catalog number: 9161028 )
- Microscope glass slides (Thermo Fisher Scientific, Menzel-Gläser, catalog number: 9161145 )
- CCD camera (Leica Microsystems, model: DC500 )
- Microscope: Nikon Eclipse TE-2000-E fluorescence microscope or Zeiss Inverted Tissue Culture Fluorescence Microscope.
Software
- Software Angio Tool (Zudaire et al., 2011) (https://ccrod.cancer.gov/confluence/display/ROB2/Home) or ImageJ software (http://imagej.nih.gov/ij/)
Procedure
- In order to visualize vessels and blood flow (leakage of blood as example), perfuse the animal for 10 min by intravenous (IV) injection (200 μl/25 gr of animal) of 20 mg/ml FITC-dextran (2,000 kDa lysine fixable at 20 μg/μl in saline). (Animals should not be anesthetized because anesthetics cause reduced heart rate and blood pressure, requiring longer perfusion times).
- Euthanize the animal by using CO2 10 min after IV injection. Enucleate the mouse eye by pulling apart the eyelids, and placing a curved forcep under the eyeball. Close the forcep and grip the connective tissue and optic nerve, being careful to avoid squeezing the eyeball, and gently pull the eyeball from the orbit. Place the eye in 10% Formalin (or 4% PFA) overnight at 4 °C to fix the tissue.
- Wash eyes after formalin-fixation 3 times for 15 min with shaking at room temperature in PBS.
- After washing the fixed, enucleated eyes, remove the cornea, iris, lens, sclera, and choroid (primary cut around the limbus). Use an 18G needle to make a small hole in the posterior of the limbus. After puncturing the limbus, use micro-scissors to cut around the circumference of the limbus. After finishing the cut, use forceps to remove the cornea, iris, lens, and sclera. Extract the vitreous gel using micro-forceps, or by gently squeezing the. In order to free the retina use two pairs of micro forceps and go around the edges to separate the retina from the eyecup and then gently squeeze the retina out. If necessary, remove the hyaloid vessels with micro forceps (Figure 1).
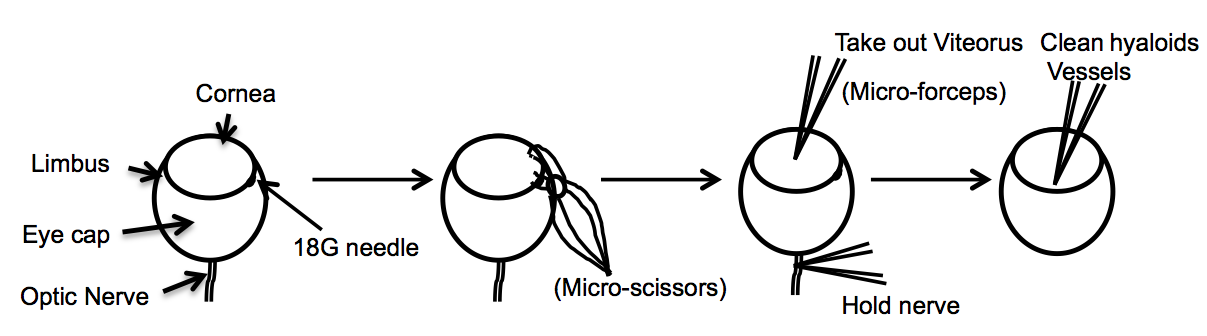
Figure 1. Diagram describes the dissection of the eye resulting in a whole mount of the retina - Incubate tissue (retinas) with blocking buffer (materials and reagents) for 1-3 h.
- Incubate retinas overnight at 4 °C under gentle agitation in blocking buffer containing BS-1 isolectin (LEC)-TRITC [incubation longer than overnight (12-24 h) may increase nonspecific fluorescence].
- Wash retinas 4-5 times (1 h between washes) in PBS at room temperature on a shaker.
- In order to flatten and mount the retina under a coverslip, make four incisions as shown in Figure 2.
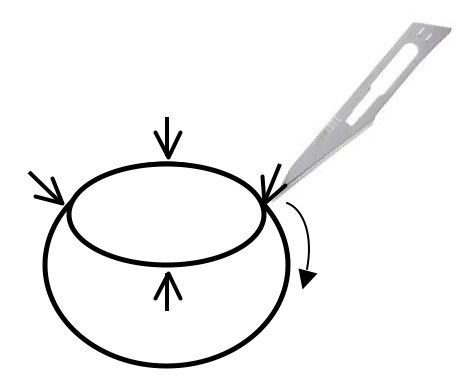
Figure 2. Retina cup is cut with surgical blade in order to allow the retinas to lie flat on the slide - Place the retina onto a slide with the eyecup facing up, and cut the retina in each quarter with a micro-scissor. Using a wide-bore plastic Pasteur pipette, remove excess PBS and spread the eyecup open like a flower. Add 1 drop (around 10 μl) of mounting media over the retina and cover with a coverslip.
- Images of flat-mounted retinal vascular plexus are acquired with an epifluorescence microscope.
To capture 3D images of the retinal vascular plexus, use a confocal microscope fitted with a CCD camera. A 5x or 10x objective offers the best image resolution for Angio Tool software. - Angio Tool image software can be used for quantifying numerous features of the angiogenic vessels including vascular density, number of branch points and tip cells, and others (Zudaire et al., 2011).
Representative data
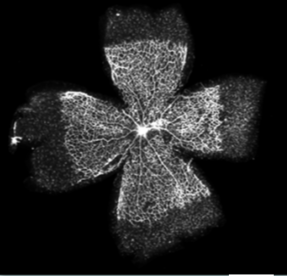
Figure 3. Representative whole mounts retina. Retinas were dissected and labeled (Alexa Fluor 594-conjugated isolectin). Scale bar: 1 mm (<5% the quality of the fluorescence-labeled are poor).
Notes
Thoroughly washing the retinas after labeling with BS-1 (LEC)-TRTIC is critical, do not reduce the washing step.
Recipes
- Blocking buffer
PBS containing 0.5% triton X-100, 0.02% sodium azide, 10% goat serum
Acknowledgments
This work was adapted and modified from previous work supported by US Public Health Service NIH grant HL071049 and part by a grant from the NIH (R01EY012726).
References
- Adini, I., Rabinovitz, I., Sun, J. F., Prendergast, G. C. and Benjamin, L. E. (2003). RhoB controls Akt trafficking and stage-specific survival of endothelial cells during vascular development. Genes Dev 17(21): 2721-2732.
- Carmeliet, P. and Jain, R. K. (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature 473(7347): 298-307.
- Stahl, A., Connor, K. M., Sapieha, P., Chen, J., Dennison, R. J., Krah, N. M., Seaward, M. R., Willett, K. L., Aderman, C. M. and Guerin, K. I. (2010). The mouse retina as an angiogenesis model. Invest Ophth Vis Sci 51(6): 2813-2826.
- Zudaire, E., Gambardella, L., Kurcz, C. and Vermeren, S. (2011). A computational tool for quantitative analysis of vascular networks. PLoS One 6(11): e27385.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Adini, I. and Ghosh, K. (2015). Mouse Retinal Whole Mounts and Quantification of Vasculature Protocol. Bio-protocol 5(15): e1546. DOI: 10.21769/BioProtoc.1546.
Category
Developmental Biology > Cell growth and fate > Angiogenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










