- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression and Purification of the Arabidopsis E4 SUMO Ligases PIAL1 and PIAL2
Published: Vol 5, Iss 15, Aug 5, 2015 DOI: 10.21769/BioProtoc.1544 Views: 7503
Reviewed by: Arsalan DaudiTzvetina BrumbarovaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2870 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1990 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
The proteins PIAL1 (At1g08910) and PIAL2 (At5g41580) are members of the recently discovered group of plant E4 SUMO ligases. This protocol allows quick and simple expression of the recombinant proteins in Escherichia coli (E. coli) and subsequent affinity purification using a maltose binding protein (MBP) tag. The proteins can be used in SUMOylation reactions, where the MBP part of the protein can be detected with a commercially available antibody, or additional purification steps can be applied.
Keywords: Small ubiquitin-related modifier SUMOMaterials and Reagents
- Rosetta DE3 pLysS strain (Merck KGaA, catalog number: 70956 )
- Vector pMAL-c2X (New England Biolabs, catalog number: N8076S ; the current version of the vector sold by NEB is pMAL-c5X, catalog number: N8108S )
- Peptone (ForMedium, catalog number: PEP03 )
- Yeast extract (ForMedium, catalog number: YEM03 )
- NaCl (Sigma-Aldrich, catalog number: S3014 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- Ampicillin (Sigma-Aldrich, catalog number: A9518 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (GERBU Biotechnik GmbH, catalog number: 1010 )
- KCl (Sigma-Aldrich, catalog number: P5405 )
- Na2HPO4 (Sigma-Aldrich, catalog number: 71505 )
- KH2PO4 (Sigma-Aldrich, catalog number: P3786 )
- Aprotinin (Sigma-Aldrich, catalog number: A6279 ) (solution stored at 4 °C)
- Leupeptin (Sigma-Aldrich, catalog number: L2884 ) (stock solution of 1 mg/ml stored in aliquots at -20 °C)
- Amylose resin (New England Biolabs, catalog number: E8021S )
- DNase I (Roche Diagnostics, catalog number: 10104159001 )
- Glycerol (GERBU Biotechnik GmbH, catalog number: 2006 )
- Ethylenediamine tetraacetic acid (EDTA) (GERBU Biotechnik GmbH, catalog number: 1034 )
- Tris (GERBU Biotechnik GmbH, catalog number: 1018 )
- Maltose (Merck KGaA, catalog number: 105911 )
- Ultrapure water (18.2 MΩ)
- Lysogeny broth (LB) (see Recipes)
- Phosphate buffered saline (PBS) (see Recipes)
- Column buffer (see Recipes)
- Elution buffer (see Recipes)
Equipment
- Orbital shaker at 37 °C
- Spectrophotometer
- Cooled centrifuge
- Bandelin Sonoplus sonicator with an MS 73 tapered probe (BANDELIN electronic GmbH & Co., model: GM70HD )
- PolyPrep Chromatography Columns (Bio-Rad Laboratories, AbD Serotec®, catalog number: 7311550 )
Procedure
- Expression
- The protein coding sequence for the E4 ligases was obtained from cDNA and inserted between the BamHI and SalI restriction sites in the vector pMAL-c2 as described in Tomanov et al. (2014). Thus, the sequence formed a continuous reading frame which was to be expressed as an N-terminal fusion to MBP (maltose binding protein).
- The plasmids were heat-shock transformed into the Rosetta DE3 pLysS strain.
- A culture of 5 ml LB with 15 mg/L chloramphenicol and 100 mg/L ampicillin was inoculated and grown overnight at 37 °C with shaking at 200 rpm.
- On the next morning, 500 µl from the overnight culture were inoculated into 200 ml fresh LB with the same antibiotics.
- The culture was incubated at 37 °C with shaking at 200 rpm until OD600 reached 0.6 (around 3 h).
- A 500 µl sample was withdrawn for electrophoretic analysis and stored at 4 °C. The culture was induced by adding IPTG to a final concentration of 1 mM.
- After three hours at 37 °C, another 500 µl sample was withdrawn and stored at 4 °C. The culture was harvested by centrifuging it at 4 °C and 4,500 x g for 20 min.
- The pellet was resuspended in PBS (pH 7.4) and centrifuged again at 4 °C and 4,500 x g for 20 min.
- The pellet was frozen at -20 °C overnight.
- The protein coding sequence for the E4 ligases was obtained from cDNA and inserted between the BamHI and SalI restriction sites in the vector pMAL-c2 as described in Tomanov et al. (2014). Thus, the sequence formed a continuous reading frame which was to be expressed as an N-terminal fusion to MBP (maltose binding protein).
- Purification
- The frozen pellet was thawed on ice.
- Five milliliters of pre-cooled column buffer were added, together with aprotinin and leupeptin at a final concentration of 1 µg/ml.
- After 15 min on ice, the bacteria were lysed using a Bandelin Sonoplus HD70 sonicator with an MS 73 tapered probe (on ice). The ultrasound bursts were applied three times for 30 sec with intermediate cooling on ice and had 60% intensity, 50% on-time.
- The suspension was centrifuged for 20 min at 4,300 x g, 4 °C.
- During the centrifugation, a BioRad PolyPrep Chromatography column (2 ml bed volume) was prepared by adding 200 µl amylose resin.
- The resin was washed with 3 column volumes (cv) water and equilibrated with 6 cv column buffer. The lower end of the column was capped and the centrifugation supernatant was added, along with 10 µg DNase I. Pre-incubation with DNase I is not necessary.
- The column was sealed and incubated for 30-60 min on a rotating wheel at 4 °C.
- The flow-through was collected and the column was washed with 24 cv column buffer.
- The protein was eluted with 3 x 1 cv elution buffer and glycerol was added to the eluates to a final concentration of 20% v/v.
- The purity and concentration of the eluted protein was checked with SDS-PAGE, and the fractions were snap-frozen in liquid nitrogen and kept at -80 °C.
- The frozen pellet was thawed on ice.
Representative data
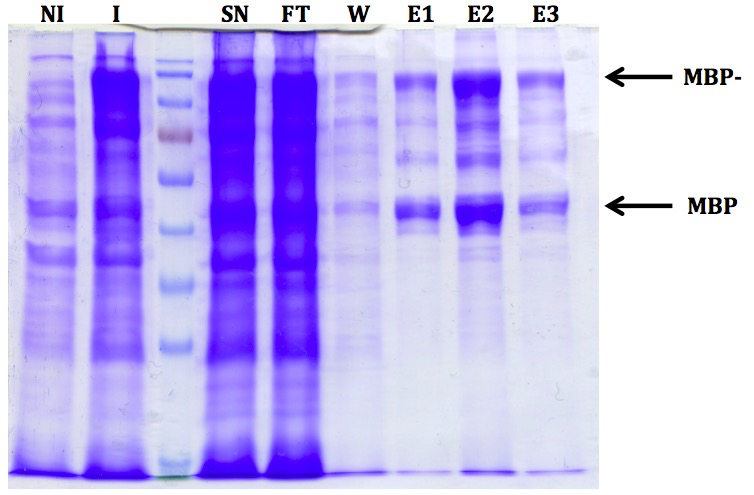
Figure 1. Representative results after expression and purification of MBP-PIAL2. Ten microliters of each fraction were mixed with 10 µl Laemmli sample buffer (Laemmli, 1970), boiled for 5 min at 95 °C and ran on a 10% SDS-PAGE gel. The non-induced (NI) and the induced (I) fraction were prepared by centrifuging 500 µl of the culture and adding 50 µl Laemmli sample buffer to the pellet. Twenty microliters were applied on the gel. Fraction names are as follows: NI - non-induced, I - induced, SN - supernatant (after sonication), FT - flow-through (after amylose resin incubation), W - wash, E - elution. Free MBP is visible in the induced fraction already and is likely a consequence of an in vivo proteolytic event.
Notes
- A more detailed explanation of SUMOylation and the enzymes involved is described by Tomanov et al. (2014).
- Judging from the usual quality of NEB products, there should not be any problems arising from using the current version of the pMAL vector.
- PBS can be prepared as a 10x stock; needs to be filtered before use.
- The column buffer can be stored for up to one month at 4 °C. The elution buffer can be stored up to six months at -20 °C. Freshly prepared buffers allow for the best results.
- Both protein fusions (MBP-PIAL1 and MBP-PIAL2) are soluble and retain their activity for in vitro SUMOylation for several months when stored at -80 °C. The pMAL-c2X vector contains a factor Xa cleavage site, so the protein can be additionally purified for crystallography studies by an in-column digest.
- For Western blot detection, NEB offers an MBP-specific antibody with catalog number E8032S.
Recipes
- Lysogeny broth (LB) (autoclaved)
1 % w/v peptone
0.5 % w/v NaCl
0.5 % w/v yeast extract
pH 7.2 - Phosphate buffered saline (PBS) (sterile filtered)
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
2 mM KH2PO4
pH 7.4 - Column buffer (sterile filtered)
20 mM Tris
200 mM NaCl
1 mM EDTA
pH 7.4 - Elution buffer (sterile filtered)
20 mM Tris
200 mM NaCl
1 mM EDTA
10 mM maltose
pH 7.4
Acknowledgments
Work in A.B.´s laboratory was supported by the Austrian Science Fund FWF (grant P25488-B22).
References
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
- Tomanov, K., Zeschmann, A., Hermkes, R., Eifler, K., Ziba, I., Grieco, M., Novatchkova, M., Hofmann, K., Hesse, H. and Bachmair, A. (2014). Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26(11): 4547-4560.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tomanov, K. and Bachmair, A. (2015). Expression and Purification of the Arabidopsis E4 SUMO Ligases PIAL1 and PIAL2. Bio-protocol 5(15): e1544. DOI: 10.21769/BioProtoc.1544.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Expression
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link