- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Nanofluidic Proteomic Immunoassay
Published: Vol 5, Iss 14, Jul 20, 2015 DOI: 10.21769/BioProtoc.1537 Views: 8939
Reviewed by: Pia GiovannelliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Phos-tag Immunoblot Analysis for Detecting IRF5 Phosphorylation
Go R. Sato [...] Tomohiko Tamura
May 20, 2017 15417 Views

Novel Protein-oligonucleotide Conjugation Method Involving a High-affinity Capture HaloTag
Junshi Yazaki
Sep 20, 2020 5556 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3421 Views
Abstract
Nanofluidic proteomic immunoassay (NIA), consisting of isoelectric focusing followed by sensitive chemiluminescence detection, has the potential to quantitatively characterize protein post-translational modifications that cause shifts in isoelectric point (pI). This protocol details the NIA analysis of protein phosphorylation using AKT as an example.
This protocol can be used for two platforms, NanoPro 1000 and Peggy Sue, from ProteinSimple Company. NanoPro 1000 separates proteins based on charge. Peggy Sue separates proteins based on either charge or size. The platforms can analyze up to 96 samples at a time with lysates from as few as 25 cells per assay. Detailed information of the platforms is available on ProteinSimple’s website (http://www.proteinsimple.com).
Materials and Reagents
- Reagents from ProteinSimple
- 25x aqueous protease/phosphatase inhibitor mix (ProteinSimple, catalog number: 040482 )
- 50x DMSO protease/phosphatase inhibitor mix (ProteinSimple, catalog number: 040510 )
- Premix G2 (Servalyt pH 5-8 separation gradient) (ProteinSimple, catalog number: 040974 )
- Premix G2 (pH 5-6 separation gradient) (ProteinSimple, catalog number: 040971 )
- PI standard 5.5 (ProteinSimple, catalog number: 040028 )
- PI standard ladder 3 (ProteinSimple, catalog number: 040646 )
- Anolyte refill (ProteinSimple, catalog number: 040337 )
- Catholyte refill (ProteinSimple, catalog number: 040338 )
- Wash concentrate refill (ProteinSimple, catalog number: 041108 )
- Amplified rabbit secondary antibody detection kit (ProteinSimple, catalog number: 041126 )
- Peroxide XDR (ProteinSimple, catalog number: 041084 )
- Amplified rabbit secondary antibody detection kit (includes SA-HRP conjugate, secondary goat-anti-rabbit-biotin conjugate and antibody diluent) (ProteinSimple, catalog number: 041126 )
- RIPA lysis buffer (ProteinSimple, catalog number: 040483 ) (see Recipes)
- Bicine/CHAPS lysis buffer and sample diluent (ProteinSimple, catalog number: 040764 ) (see Recipes)
- Anolyte (10 mM Phosphoric Acid) (see Recipes)
- Catholyte (100 mM Sodium Hydroxide) (see Recipes)
- 25x aqueous protease/phosphatase inhibitor mix (ProteinSimple, catalog number: 040482 )
- Antibodies for AKT analysis
- AKT (pan) antibody (Cell Signaling Technology, catalog number: 4691 )
- AKT1 antibody (BD Biosciences, catalog number: 610861 )
- AKT2 antibody (Cell Signaling Technology, catalog number: 3063 )
- Phospho-AKT (Thr308) (Cell Signaling Technology, catalog number: 2965 )
- Phospho-AKT (Thr450) (Cell Signaling Technology, catalog number: 9267 )
- Phospho-AKT (Ser473) (Cell Signaling Technology, catalog number: 9271 )
- AKT (pan) antibody (Cell Signaling Technology, catalog number: 4691 )
- Other materials
- BCA protein assay kit (Pierce Antibodies, catalog number: 23225 )
- Dulbecco’s Modification of Eagle’s Medium (DMEM) with 4.5 g/L glucose & L-glutamine without pyruvate (Mediatech, catalog number: 10017CV )
- McCoy’s 5A (1x, with L-glutamine) (Mediatech, catalog number: 10050CV )
- Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10438026 )
- BCA protein assay kit (Pierce Antibodies, catalog number: 23225 )
Equipment
- NP1000 or Peggy (ProteinSimple, model: 004800 )
- Centrifuge for microplates (Thermo Fisher Scientific, model: Sorvall Legend XTR )
- Capillaries Charge Separation (ProteinSimple, catalog number: CBS701 )
- Assay Plate/Lid Kit (ProteinSimple, catalog number: 040663 )
- Sponge Pack (ProteinSimple, catalog number: 041528 )
- Tissue culture dish (100 mm) (Corning Incorporated, catalog number: 430167 )
- Microcentrifuge tube (Denville Scientific, catalog number: C2170 )
Software
- Compass version 2.5.11 (ProteinSimple)
Procedure
- Cell culture and lysate preparation
- Culture cells (such as HeLa or HCT116 cells) in DMEM or McCoy’s 5A medium supplemented with 10% FBS to 80-90% confluence in 10 cm plates. Treat cells as required.
- Gently wash cells in plates with 10 ml ice-cold PBS. Aspirate PBS thoroughly.
- Add 400 μl ice-cold RIPA or Bicine/CHAPS lysis buffer supplemented with both aqueous and DMSO protease/phosphatase inhibitor mix to each 10 cm plate on ice. Gently swirl plates to ensure good coverage and incubate for 10 min on ice.
- Scrape cells thoroughly. Pipet up and down to mix. Transfer buffer and cells to a 1.5 ml microfuge tube and incubate for additional 20 min on ice. Vortex the tube briefly every 5 min.
- Clarify by centrifugation (14,000 x g, 10 min) at 4 °C.
- Transfer supernatant to a fresh microfuge tube. Immediately aliquot supernatant (10-30 μl per aliquot) on ice and store at -80 °C.
- Measure protein concentrations using the BCA protein assay kit.
- Culture cells (such as HeLa or HCT116 cells) in DMEM or McCoy’s 5A medium supplemented with 10% FBS to 80-90% confluence in 10 cm plates. Treat cells as required.
- Sample preparation for loading (for 12 lysates)
- In a microfuge tube, combine 110 µl premix G2 pH5-6, 110 µl premix G2 Servalyte pH5-8, 3.95 µl ladder 3, 1 µl Standard 5.5, and 4.5 µl DMSO protease/phosphatase inhibitor.
- Vortex for at least 15 sec to mix thoroughly. Aliquot 19 µl of the mix to each of 12 microfuge tubes.
- Dilute 5 μg of high concentration lysate with sample diluent or Bicine/CHAPS buffer to a final volume of 6 µl.
- Add the 6 µl of diluted lysate into one of the 12 tubes with 19 µl of the mix. The final protein concentration in capillary is 0.2 µg/µl.
- Add 10 µl of lysate mix into wells in row A (see Table 2).
- In a microfuge tube, combine 110 µl premix G2 pH5-6, 110 µl premix G2 Servalyte pH5-8, 3.95 µl ladder 3, 1 µl Standard 5.5, and 4.5 µl DMSO protease/phosphatase inhibitor.
- Primary antibody preparation
- Prepare primary antibodies at 1:50 dilution with antibody diluent (from the secondary antibody detection kit) in microfuge tubes.
- Add 10-20 μl of diluted primary antibodies into appropriate wells of 384-well assay plate (see Table 2).
- Prepare primary antibodies at 1:50 dilution with antibody diluent (from the secondary antibody detection kit) in microfuge tubes.
- Secondary antibody preparation
- Prepare secondary antibodies at 1:100 dilution with antibody diluent in microfuge tubes.
- Add 10-20 μl of diluted secondary antibodies into appropriate wells of 384-well assay plate (see Table 2).
- Prepare secondary antibodies at 1:100 dilution with antibody diluent in microfuge tubes.
- Streptavidin-HRP preparation
- Prepare SA-HRP at 1:100 dilution with antibody diluent.
- Add 10-20 μl of diluted SA-HRP into appropriate wells of 384-well assay plate (see Table 2).
- Prepare SA-HRP at 1:100 dilution with antibody diluent.
- Peroxide/Luminol preparation
Combine 120 µl XDR peroxide and 120 µl luminol, mix well, and then add 10-20 μl of the mix into appropriate wells of the assay plate (see Table 2). - Centrifuge the assay plate at 2,500 x g for 5 min to spin down the liquid and eliminate bubbles. Protect the plate from light before loading into the equipment.
- While the plate is spinning, create the assay file in Compass software. Set up as below for AKT assay. Table 1. Parameters of instrument settings
Instrument setting Parameters Sample loading time 25 sec Separation conditions 40,000 μW, 40 min UV immobilization time 60 sec Wash 1 2 washes, 150 sec each (default) Primary antibody incubation 120 min (default) Wash 2 2 washes, 150 sec each (default) Secondary antibody incubation 60 min (default) Wash 3 2 washes, 150 sec each (default) Chemiluminescence exposure time 30, 60, 120, 240 and 480 sec
Table 2. An example of an assay layout. Assays are set up in 384-well plates. Table 2 was an example of an assay layout for 12 samples and 6 antibodies. Lysates were added into wells in row A from A1 to A12, respectively. Diluted primary antibodies were added into wells in row B (B1 to B12) to row G. Diluted secondary antibodies were added in row H (H1 to H12). Streptavidin-HRP was added into row I (I1 to I12). To avoid possible contamination between streptavidin-HRP and detection reagents in the plate, Lumino/Peroxide XDR was added into row A (A13 to A24) away from streptavidin-HRP.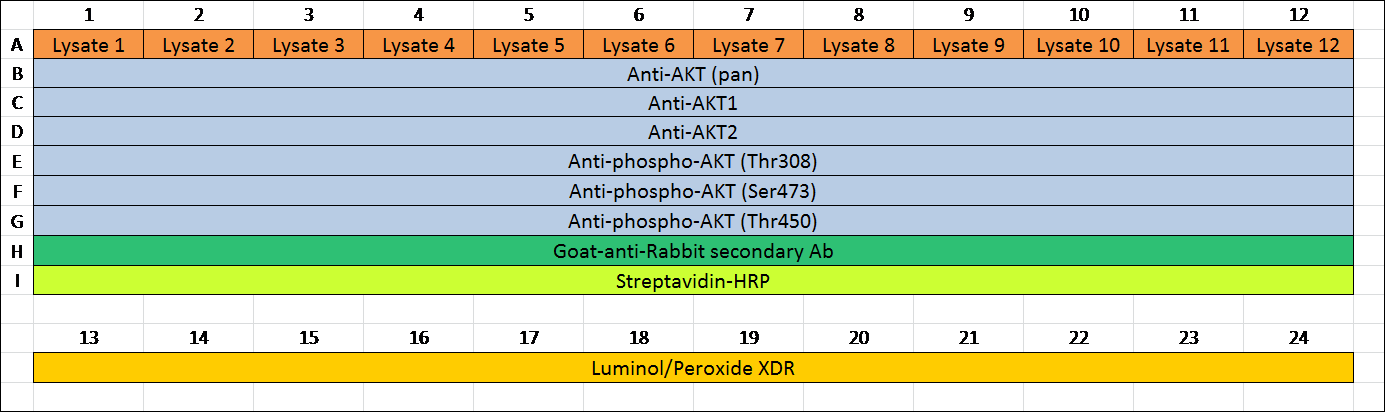
- Start the run
Launch the Start Run Wizard:- Empty waste bottle and fill wash bottle with fresh ultrapure water.
- Replace sponge.
- Fill the wash buffer, Anolyte and Catholyte cups.
- Put capillary box in the primary capillary box position.
- Load sample plate.
- Click the “RUN” button to start the assay.
- Empty waste bottle and fill wash bottle with fresh ultrapure water.
- Data analysis
Data analysis is performed with the Compass software provided by ProteinSimple.- In Image Analysis Settings, view data for different exposures in the Analysis screen. Choose the best exposure time according to the intensity of the signals.
- In Peak Fit Analysis Setting, set up Range (for AKT): minimum: 4.5, Maximum: 7.0.
- In Standard Analysis Settings, choose Std. Ladder 3 (premix pI 5-8) and add Standard 5.5 to the pI column and enter 500 in the Position column of the Fluorescent Peaks table. Select 4.9 and 7.0 for capillary registration by clicking the checkbox in the Registration column.
- In Image Analysis Settings, view data for different exposures in the Analysis screen. Choose the best exposure time according to the intensity of the signals.
- Start the run
Representative data
- Representative results of the assay:
Multiple peaks of AKT were detected by NIA using total AKT antibody representing both AKT1 and AKT2 isoforms as well as differences in post-translational modification (PTM) of each molecule (Fig. 1). No AKT3 is expressed in HCT116 cells.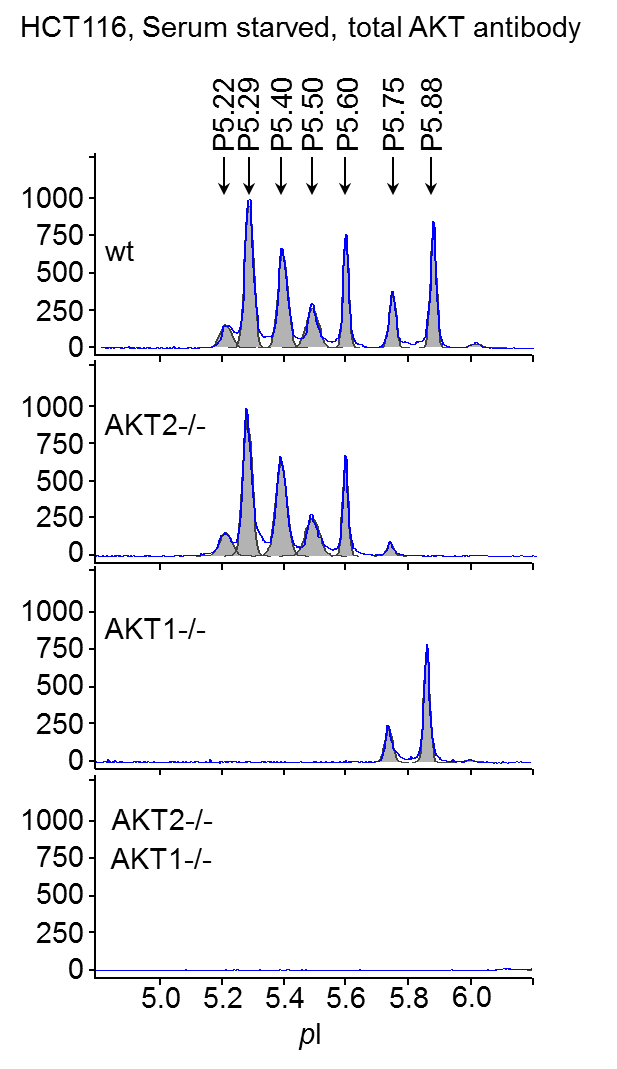
Figure 1. AKT isoforms characterized by NIA. Wt, AKT2-/-, AKT1-/-, and AKT1, 2 double knockout HCT116 cells were serum-starved overnight and lysed in NIA RIPA buffer. Samples were analyzed with anti-total AKT antibody. pI is shown on x axis and chemiluminescence on y axis.
Notes
- Keep samples on ice all the time.
- Always use protease and phosphatase inhibitors to preserve phosphorylation signals.
- Try to maintain low sample ionic strength to avoid shifts during IEF separation. The final total salt concentration should not exceed 50 mM in the capillary. Bicine/CHAPS or RIPA lysis buffers from ProteinSimple are recommended. It is imperative to obtain a high protein concentration if cells are harvested in RIPA buffer.
- Capillaries are light and humidity sensitive. Protect capillaries in opened packs from light and humidity.
Recipes
- RIPA lysis buffer
20 mM HEPES (pH 7.5)
150 mM NaCl
1 % NP40 alternative
0.25 % sodium deoxycholate - Bicine/Chaps lysis buffer
20 mM bicine (pH 7.6)
0.6% CHAPS - Anolyte (10 mM phosphoric acid)
- Catholyte (100 mM sodium hydroxide)
Acknowledgments
The work was supported by National Institutes of Health (NIH) grant 5R21CA126700 and a grant from the University of Texas MD Anderson Cancer Center Kidney Cancer Multidisciplinary Research Program to ZD. The protocol was adapted from our published paper in Oncogene (Guo et al., 2014) with modifications.
References
- Compass Software for NanoPro 1000 User Guide. (P/N 001-506, Revision 2, June 2013)
- Guo, H., Gao, M., Lu, Y., Liang, J., Lorenzi, P. L., Bai, S., Hawke, D. H., Li, J., Dogruluk, T., Scott, K. L., Jonasch, E., Mills, G. B. and Ding, Z. (2014). Coordinate phosphorylation of multiple residues on single AKT1 and AKT2 molecules. Oncogene 33(26): 3463-3472.
- ProteinSimple Nanopro Assay Development Guide. (P/N 041-015, Revision 3a, July 2011)
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Guo, H., Lorenzi, P. L. and Ding, Z. (2015). Nanofluidic Proteomic Immunoassay. Bio-protocol 5(14): e1537. DOI: 10.21769/BioProtoc.1537.
Category
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









