- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Simple Method to Generate Gene Knockout Clones in Human Cells Using Transcription Activator-Like Effector Nuclease (TALEN)
Published: Vol 5, Iss 14, Jul 20, 2015 DOI: 10.21769/BioProtoc.1531 Views: 10172
Reviewed by: Pinchas TsukermanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Generation of Gene Knockout and Gene Replacement with Complete Removal of Full-length Endogenous Transcript Using CRISPR-Trap
Jonas Mechtersheimer [...] Marc-David Ruepp
Oct 20, 2018 9390 Views

In vitro Generation of CRISPR-Cas9 Complexes with Covalently Bound Repair Templates for Genome Editing in Mammalian Cells
Nataša Savić [...] Gerald Schwank
Jan 5, 2019 11889 Views

GeneWeld: Efficient Targeted Integration Directed by Short Homology in Zebrafish
Jordan M. Welker [...] Maura McGrail
Jul 20, 2021 11733 Views
Abstract
Transcription activator-like effectors (TALEs) are naturally occurring proteins secreted by the plant pathogen, Xanthomonas, and fused to the Fok1 endonuclease to generate TALE nucleases (TALENs). TALEN pairs bind to specific DNA sequences initiating Fok1 dimerization and double-stand cleavage of DNA within the TALEN target site. This cleavage event triggers cellular repair mechanisms that result in insertions and/or deletions (indels), which enable gene knockout. The high specificity and efficiency of TALENs makes them important tools for genome editing. Here, we describe a method for the generation of single-cell clones with targeted gene knockout by TALEN using co-transfection and FACS with a fluorescent reporter. This protocol was designed to knockout cell death-inducing DFFA-like effector b, CIDEB, in Huh7.5 cells; however, this protocol can be applied to a wide range of cell types and genes of interest.
Materials and Reagents
- Huh7.5 cells (Charles Rice lab, Rockefeller University)
- Dulbecco’s modified Eagle’s medium (DMEM high glucose) (HyClone, catalog number: SH30243.02 )
- Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10437-028 )
- Antibiotic Antimycotic solution (10,000 U/ml penicillin G, 10,000 µg/ml streptomycin, 25 µg/ml amphotericin B) (HyClone, catalog number: SV30079.01 )
- MEM nonessential amino acids (NEAA) (Corning Incorporated, catalog number: 25025CI )
- Phosphate buffered saline (PBS) (Thermo Fisher Scientific, catalog number: BP39920 )
- Trypsin, 0.05% (HyClone, catalog number: SH30236.02 )
- TALEN Sure KO plasmids (Cellectis)
Note: It is custom designed to target the gene of interest and includes right and left TALEN plasmids to bind both sequences of DNA flanking the target site. - pcDNA3-EGFP expression plasmid (Addgene, plasmid number: 13031 )
- Lipofectamine 2000 Transfection Reagent (Life Technologies, InvitrogenTM, catalog number: 11668019 )
- Opti-MEM I Reduced-Serum Medium (Life Technologies, Gibco®, catalog number: 31985070 )
- QuickExtract DNA Extraction Solution (Epicentre, catalog number: QE09050 )
- Platinum PCR SuperMix High Fidelity (Life Technologies, InvitrogenTM, catalog number: 12532016 )
- Primers flanking the TALEN target site (custom DNA nucleotides) (Integrated DNA Technologies)
- QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28704 )
- Tris-HCl buffer (pH 6.8, 0.5 M) (Bio-Rad Laboratories, AbD Serotec®, catalog number: 1610799 )
- Sodium dodecyl sulfate (20% solution) (Thermo Fisher Scientific, catalog number: BP13111 )
- Glycerol (Fisher Scientific, catalog number: BP2291 )
- Bromophenol blue (Thermo Fisher Scientific, catalog number: B3925 )
- 2-mercaptoethanol (Thermo Fisher Scientific, catalog number: O3446I100 )
- 10% DMEM culture media (see Recipes)
- 20% DMEM culture media (see Recipes)
- FACS buffer (see Recipes)
- 2x SDS loading buffer (see Recipes)
Equipment
- Cell culture incubator at 37 °C and 5% CO2
- Cell culture-treated polystyrene plates (100 mm, 6 well, 12 well, 24 well, 48 well, 96 well) (Corning Incorporated)
- Polystyrene Round-Bottom Tube (5 ml FACS tubes) (BD Biosciences, Falcon®, catalog number: 352008 )
- Round-Bottom Tube Cap (5 ml) (BD Biosciences, Falcon®, catalog number: 352032 )
- 40 micron cell strainer (Falcon®, catalog number: 352340 )
- Fluorescence microscope
- Table top centrifuge
- FACSAria cell sorter (BD Biosciences, catalog number: 656700 )
- Multi-channel pipette
- Western blotting equipment (Bio-Rad Laboratories, AbD Serotec®)
- Heat block (VWR international)
- PCR thermocycler (Bio-Rad Laboratories, AbD Serotec®)
- Agarose gel electrophoresis system
- 3730 DNA analyzer (Life Technologies, Applied Biosystems®, catalog number: 3730S )
Procedure
- Co-transfection of TALEN and EGFP plasmids into Huh7.5 cells
- Plate Huh7.5 cells onto 100 mm cell culture plates in 15 ml 10% DMEM culture media without antibiotics 18-24 h before transfection such that the cells will be ~60-70% confluent at the time of transfection. Scale up and down the culture according to the manufacturer’s protocol for the cell type used.
- Combine 8 µg of the right TALEN, 8 µg of the left TALEN and 8 µg of EGFP plasmid in 1.5 ml Opti-MEM medium. Mix gently. Prepare separate tubes for controls, such as mock transfection, which controls for potential side effects of the transfection reagent (only to be included in step A).
- In a separate tube, add 60 µl Lipofectamine 2000 to 1.5 ml Opti-MEM medium for each sample. Mix gently and incubate at room temperature for 5 min.
- Add the DNA-Opti-MEM solution (from step A2) to the Lipofectamine-Opti-MEM solution (from step A3). Thoroughly mix the solution by gently pipetting up and down. Incubate at room temperature for 20 min.
- Add 1.5 ml of the DNA-lipid complex to the cells drop-wise and gently rock the plate back and forth to evenly distribute the complexes.
- (Optional) ~6 h later, replace the media with fresh 10% DMEM culture media.
- Incubate the cells at 37 °C with 5% CO2 for 48 h.
- Approximate transfection efficiency by measuring the number of GFP positive cells using a fluorescence microscope.
- Plate Huh7.5 cells onto 100 mm cell culture plates in 15 ml 10% DMEM culture media without antibiotics 18-24 h before transfection such that the cells will be ~60-70% confluent at the time of transfection. Scale up and down the culture according to the manufacturer’s protocol for the cell type used.
- Enriching for GFP positive cells using fluorescence activated cell sorting (FACS)
- 48 h post-transfection, wash cells with 1x PBS, then trypsinize cells and pipette up and down forcefully to break up cell aggregates.
- Add 10% DMEM culture media to inactivate trypsin.
- Centrifuge cells at 200 x g for 10 min.
- Aspirate supernatant and resuspend the cell pellet in FACS buffer (see Recipes) at a concentration of 10 x 106 cells/ml. Pipette up and down thoroughly to attain single cell suspensions.
- Place a cell strainer over a FACS tube (sterilized in ethanol) and carefully pipet the cells through the filter into the tube to remove cell clumps. Cells should be sorted immediately or placed on ice and covered until ready to be sorted.
- Sort the GFP positive cells into 20% DMEM culture media using the FACSAria cell sorter.
- Cell viability is typically diminished following transfection and sorting. Thus, the sorted cells should be seeded at a fairly high cell density in fresh 20% DMEM culture media and allowed to recover for ~1 week. Replace the media every 3-4 days during this recovery time.
Note: If cells fail to recover, repeat the experiment starting with a larger cell number.
- 48 h post-transfection, wash cells with 1x PBS, then trypsinize cells and pipette up and down forcefully to break up cell aggregates.
- Generation of single-cell clones by serial dilution onto 96 well plates
- Once the cells have recovered, add 100 µl of 20% DMEM culture media to all but the first well, A1, of a 96 well plate.
- Suspend sorted cells at a concentration of 1 x 104 cells/ml.
- Add 200 µl of sorted cell suspension to well A1, then transfer 100 µl to well B1, and so on, until all the wells in the first column contain cells. Discard 100 µl from H1, and then add 100 µl of media to wells A1-H1 for a final volume of 200 µl.
- Using a multi-channel pipette, transfer 100 µl from the first column to the second column and repeat for all columns. Bring the final volume of all wells to 200 µl.
- Incubate cells at 37 °C with 5% CO2 in 20% DMEM culture media. Analyze the wells daily marking those that contain a single cell. Close monitoring and stringency is crucial at this step for obtaining single-cell clones.
- Allow single-cell clones to become confluent in 96 well plates, then gently trypsinize and transfer to 48 well plates. Repeat this process so that the cells are slowly expanded to larger surface areas to promote cell growth (e.g. transfer from 96 well to 48 well to 24 well to 12 well to 6 well plates).
- Once the cells have recovered, add 100 µl of 20% DMEM culture media to all but the first well, A1, of a 96 well plate.
- Analyze protein expression by Western blotting to identify single-cell clones with gene knockout
- In 12 well plates, wash cells with 1x PBS then collect cells by trypsinization. Add 10% DMEM culture media to inactivate trypsin.
- Centrifuge cells at 200 x g for 10 min.
- Aspirate supernatant and lyse the cells with 2x SDS loading buffer (see Recipes) to extract protein.
- Immediately boil samples at 100 °C for 10 min using a hotplate, then centrifuge at max speed for 1 min. Samples can be stored at -20 °C.
- Analyze protein expression via Western blotting (Figure 1A).
- In 12 well plates, wash cells with 1x PBS then collect cells by trypsinization. Add 10% DMEM culture media to inactivate trypsin.
- Confirm gene knockout by DNA sequence analysis
- Extract DNA using QuickExtract DNA solution per the manufacturer’s protocol.
- Perform PCR with genomic primers flanking the TALEN target site.
- Analyze the PCR products by gel electrophoresis.
- Isolate and purify DNA using the Qiagen gel extraction kit per the manufacturer’s protocol.
- Sequence DNA to confirm the presence of TALEN-induced indels that result in the loss of gene expression (Figure 1B).
- Extract DNA using QuickExtract DNA solution per the manufacturer’s protocol.
Representative data
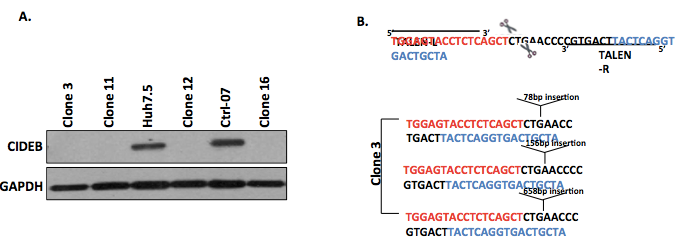
Figure 1. Representative data. A. Representative Western blot for the identification of 4 clones negative for CIDEB protein expression, suggesting successful gene knockout by TALEN. B. Representative DNA sequence analysis of clone 3 confirming that the target region contains TALEN-induced indels leading to loss of gene expression.
Notes
- Pertaining to step B, Huh7.5 cells display low transfection efficiencies (~10-15%); thus, FACS is an important step to enrich for GFP+ cells. However, sorting may not be necessary for cell lines with high transfection efficiencies such as 293T, HeLa, and RKO cells. If the transfection efficiency is determined to be over 80% by fluorescence microscopy and/ or flow cytometry, then the transfected cells from step A may be directly used for single-cell cloning in step C.
- Various protocols exist for single-cell cloning including by serial dilution (described here), by cloning rings, and by directly counting and plating cells at 0.5 cells per well. The best method will be determined by the cell line used and viability of the cells after gene knockout.
- Depending on the available equipment and size of the screen, it may be easier to analyze the clones at the DNA level first, and then confirm positive clones by Western blotting. This is at the experimenter’s discretion.
Recipes
- 10% DMEM culture media
DMEM supplemented with 10% FBS, 1x NEAA, 100 U/ml penicillin G, 100 µg/ml streptomycin and 0.25 µg/ml amphotericin B - 20% DMEM culture media
DMEM supplemented with 20% FBS, 1x NEAA, 100 U/ml penicillin G, 100 µg/ml streptomycin and 0.25 µg/ml amphotericin B - FACS buffer
PBS with 5% FBS - 2x SDS gel-loading buffer
0.1 M Tris-HCl (pH 6.8) with 4% sodium dodecyl sulfate (SDS), 20% glycerol, 0.2% bromophenol blue, and 10% β-mercaptoethanol
Acknowledgments
This work was supported by NIH grant R01 A1079150.
References
- Wu, X., Lee, E. M., Hammack, C., Robotham, J. M., Basu, M., Lang, J., Brinton, M. A. and Tang, H. (2014). Cell death-inducing DFFA-like effector b is required for hepatitis C virus entry into hepatocytes. J Virol 88(15): 8433-8444.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hammack, C. (2015). A Simple Method to Generate Gene Knockout Clones in Human Cells Using Transcription Activator-Like Effector Nuclease (TALEN). Bio-protocol 5(14): e1531. DOI: 10.21769/BioProtoc.1531.
Category
Cell Biology > Cell engineering > TALEN
Molecular Biology > DNA > Mutagenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








