- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification of Factors in Regulating a Protein Ubiquitination by Immunoprecipitation: a Case Study of TRF2 on Human REST4 Ubiquitination
Published: Vol 5, Iss 13, Jul 5, 2015 DOI: 10.21769/BioProtoc.1525 Views: 12261
Reviewed by: Vivien Jane Coulson-ThomasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Phos-tag Immunoblot Analysis for Detecting IRF5 Phosphorylation
Go R. Sato [...] Tomohiko Tamura
May 20, 2017 15417 Views

Novel Protein-oligonucleotide Conjugation Method Involving a High-affinity Capture HaloTag
Junshi Yazaki
Sep 20, 2020 5557 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3423 Views
Abstract
Ubiquitination is the first step of the ubiquitin-proteasome pathway that regulates cells for their homeostatic functions and is an enzymatic, protein post-translational modification process in which ubiquitin is transferred to a target protein substrate by a set of three ubiquitin enzymes (Weissman et al., 2011; Bhattacharyya et al., 2014; Ristic et al., 2014). Given the importance of this process, it is plausible that ubiquitination is under strict control by many factors and that the regulatory machineries are protein-specific. An assay for the detection of a specific protein ubiquitination will enable us to examine whether a factor has a function to regulate the ubiquitination of this protein. Here we describe a protocol that detects the ubiquitination status of the human REST4 protein in cultured cells, a neural alternative splicing isoform of REST (RE-1 silencing transcription factor), that antagonizes the repressive function of REST on neural differentiation and neuron formation. Using this protocol, we show that the telomere binding protein TRF2 stabilizes the expression of the human REST4 by inhibiting its ubiquitination. This indicates that TRF2 plays a positive role in neural differentiation (Ovando-Roche et al., 2014). This protocol is also useful for the detection of ubiquitination of other proteins of interest.
Keywords: TRF2Materials and Reagents
- Cells and plasmid
- TRF2-HEK293T cells: 293T cells (ATCC, catalog number: CRL-3216 ) that have been stably transfected with TRF2 expressing vector
- Control-HEK-293T cells: 293T cells (ATCC, catalog number: CRL-3216) that have been stably transfected with empty control vector
- Myc-REST4 expressing vector: generated by modifying the Myc-REST plasmid (Huang et al., 2011) as detailed in Ovando-Roche et al. (2014)
- HA-ubiquitin expressing vector: (Addgene, catalog number: 17608 )
- TRF2-HEK293T cells: 293T cells (ATCC, catalog number: CRL-3216 ) that have been stably transfected with TRF2 expressing vector
- Antibodies
- Anti-TRF2 antibody (EMD Millipore, catalog number: 05-521 )
- Anti-HA antibody (Sigma-Aldrich, catalog number: H6908 )
- Anti β-Actin antibody (Sigma-Aldrich, catalog number: A5441 )
- C-Myc antibody (Santa Cruz, catalog number: SC-40 )
- Goat anti-mouse light chain specific (Jackson ImmunoResearch Laboratories, catalog number: 115-035-174 )
- Mouse anti-rabbit light chain specific (Jackson ImmunoResearch Laboratories, catalog number: 211-032-171 )
- Normal mouse IgG (Santa Cruz, catalog number: SC-2025 )
- Anti-TRF2 antibody (EMD Millipore, catalog number: 05-521 )
- Reagents
- Dulbeccos modified eagles medium (DMEM) (Sigma-Aldrich, catalog number: D5671 )
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F7524 )
- Calcium phosphate transfection kit (Sigma-Aldrich, catalog number: CAPHOS )
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Bromophenol blue (Bio-Rad Laboratories, catalog number: 161-0404 )
- CaCl2 (VWR International, catalog number: 22317.297 )
- Dynabeads protein G (Life Technologies, catalog number: 10003D )
- EDTA (Sigma-Aldrich, catalog number: ED-1KG )
- EGTA (Sigma-Aldrich, catalog number: E4378 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- KCl (VWR International, catalog number: JT3040-1 )
- MG132 (Calbiochem, catalog number: 474791 )
- MgCl2 (Sigma-Aldrich, catalog number: 63063 )
- NaCl (Sigma-Aldrich, catalog number: s-3014 )
- NaOH (VWR International, catalog number: 28244.263 )
- Nonidet P40 (VWR International, catalog number: E109-100ML )
- Phenylmethanesulfonyl fluoride (Sigma-Aldrich, catalog number: 93482 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340 )
- SDS (Sigma-Aldrich, catalog number: 71725 )
- Tris-HCl (Sigma-Aldrich, catalog number: T3253 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Tween 20 (Sigma-Aldrich, catalog number: P9416 )
- Ubiquitin aldehyde (Boston Biochem, catalog number: U-201 )
- Dulbeccos modified eagles medium (DMEM) (Sigma-Aldrich, catalog number: D5671 )
- Buffers
- Antibody binding buffer (see Recipes)
- HEPES lysis buffer (see Recipes)
- 2x electrophoresis sample buffer (see Recipes)
- Antibody binding buffer (see Recipes)
Equipment
- 100 mm cell culture dish (Corning, catalog number: 430167 )
- Cell scraper (Corning, catalog number: 3010 )
- 1.5 ml Eppendorf tubes (StarLab, catalog number: S1615-5500 )
- BD Microlance 3 21G needle (BD, catalog number: 301156 )
- 37 °C, 5% CO2 forced-air incubator (Binder, model: C150 )
- Microcentrifuge (Hettich, model: 200R )
- Thermomixer compact (Eppendorf, model: 5350 )
- Rotator (Stuart, model: SB3 )
- Dynamag 2 magnet (Life Technologies, catalog number: 12321D )
Procedure
- Culture TRF2-HEK293T and control cells in 100 mm dishes with a standard medium (DMEM containing 10% FBS). Upon reaching 60-70% confluency, co-transfect the cells with plasmids Myc-REST4/HA-Ubiquitin using Calcium phosphate transfection kit following manufacturer’s instruction.
- 48 h after the transfection, treat the cells with 20 µM MG132 (protect from light) in the standard medium and incubate for 5 h at 37 °C, 5% CO2.
- Remove medium and wash cells with PBS, then lyse cells in a 100 mm dish with 600 µl of chilled HEPES lysis buffer (incubate on ice/4 °C for 4 min before scraping to increase protein yield).
- Collect cell lysate in an Eppendorf tube and pass lysate through 21G needle ~25 times to aid lysis and shred DNA.
- Add ubiquitin aldehyde to the lysate to a final concentration of 1 µM and mix sample on a rotator at 20 rpm for 15 min at 4 °C.
- Centrifuge the sample at 12,000 x g for 15 min at 4 °C to pellet cell debris and transfer the supernatant to a new tube.
- Pre-clear samples with 50 µl of protein G Dynabeads for 1 h at 4 °C with a rotator at 20 rpm to remove proteins that non-specifically stick to beads.
- Bind antibodies to beads for immunoprecipitation (IP):
- Vortex Dynabeads stock for >30 sec to re-suspend beads.
- Dispense 50 µl of beads per IP reaction.
- Place on magnet to separate beads from solution, aspirate out solution.
- Dilute 3 µg Myc antibody in 500 µl of binding buffer and do the same for normal mouse IgG control.
Note: Since Myc antibody was raised from mouse, normal mouse IgG is used as negative control. - Bind antibodies to the beads by incubating 30 min at room temperature and 30 min at 4 °C with shaking.
- Vortex Dynabeads stock for >30 sec to re-suspend beads.
- Quantify the pre-cleared sample for protein concentration and prepare 1 mg protein per IP and 20 µg protein per input.
- After antibody-beads binding, place tube containing beads onto magnet and remove binding solution. Wash beads once with binding solution to remove any excess antibody.
- Transfer 1 mg of pre-cleared proteins into the beads that have been linked to Myc antibody or normal mouse IgG. Mix samples and incubate for 2 h at 4 °C with rotation at 20 rpm to perform IP.
- Following formation of the beads-antibody-antigen complex, place samples back onto magnet to remove the supernatant.
- Re-suspend beads in 400 µl lysis buffer and place back onto magnet, remove supernatant as before.
- Repeat step 13 two more times to wash beads of any unspecific binding.
- After final wash re-suspend beads-antibody-antigen complex in 100 µl lysis buffer and transfer to a clean tube.
- Place the samples in magnet and remove the supernatant. Re-suspend complex pellet in 40 µl 2x electrophoresis buffer, vortex harshly and pulse spin in microcentrifuge to collect all the solution to the bottom of the tube.
- Heat the above sample at 70 °C for 10 min with shaking in a thermomixer.
- Place the sample back onto magnet for a final time and transfer supernatant to a new tube (IP tube).
- Boil the IP samples and Input samples (20 µg of lysate directly from step 9 for each sample) at 95 °C for 5 min.
- Analyze the samples via Western blotting.
- Specificity and quality of IP: In order to verify the function of TRF2 on REST4 ubiquitination, it is necessary to ensure that similar amount of Myc-REST4 proteins are precipitated between TRF2 expressing and control cells and that this precipitation is specific. Therefore, IP samples are tested with anti-c-Myc (1:1,000) antibody as shown in Figure 1.
- Effect of TRF2 on REST4 ubiquitination: IP samples are also tested for ubiquitin binding with anti-HA (1:1,000 dilution) antibody as ubiquitin is tagged with HA epitope.
- To ensure the correct protein expression and loading, all cell lysates without immunoprecipitation (input samples) are tested with anti-c-Myc (1:1,000), anti-TRF2 (1:500) and anti-β-actin (1:5,000) antibodies.
- Specificity and quality of IP: In order to verify the function of TRF2 on REST4 ubiquitination, it is necessary to ensure that similar amount of Myc-REST4 proteins are precipitated between TRF2 expressing and control cells and that this precipitation is specific. Therefore, IP samples are tested with anti-c-Myc (1:1,000) antibody as shown in Figure 1.
Representative data
- Here is a representative result showing that ubiquitination of REST4 is clearly detected in the cells that were co-transfected with Myc-REST4 and HA-tagged ubiquitin (HA-Ub) and that expression of TRF2 reduces the ubiquitination of REST4 in these cells. The results suggest that TRF2 stabilizes REST4 expression by inhibition of its ubiquitin-proteasomal degradation (Ovando-Roche et al., 2014).
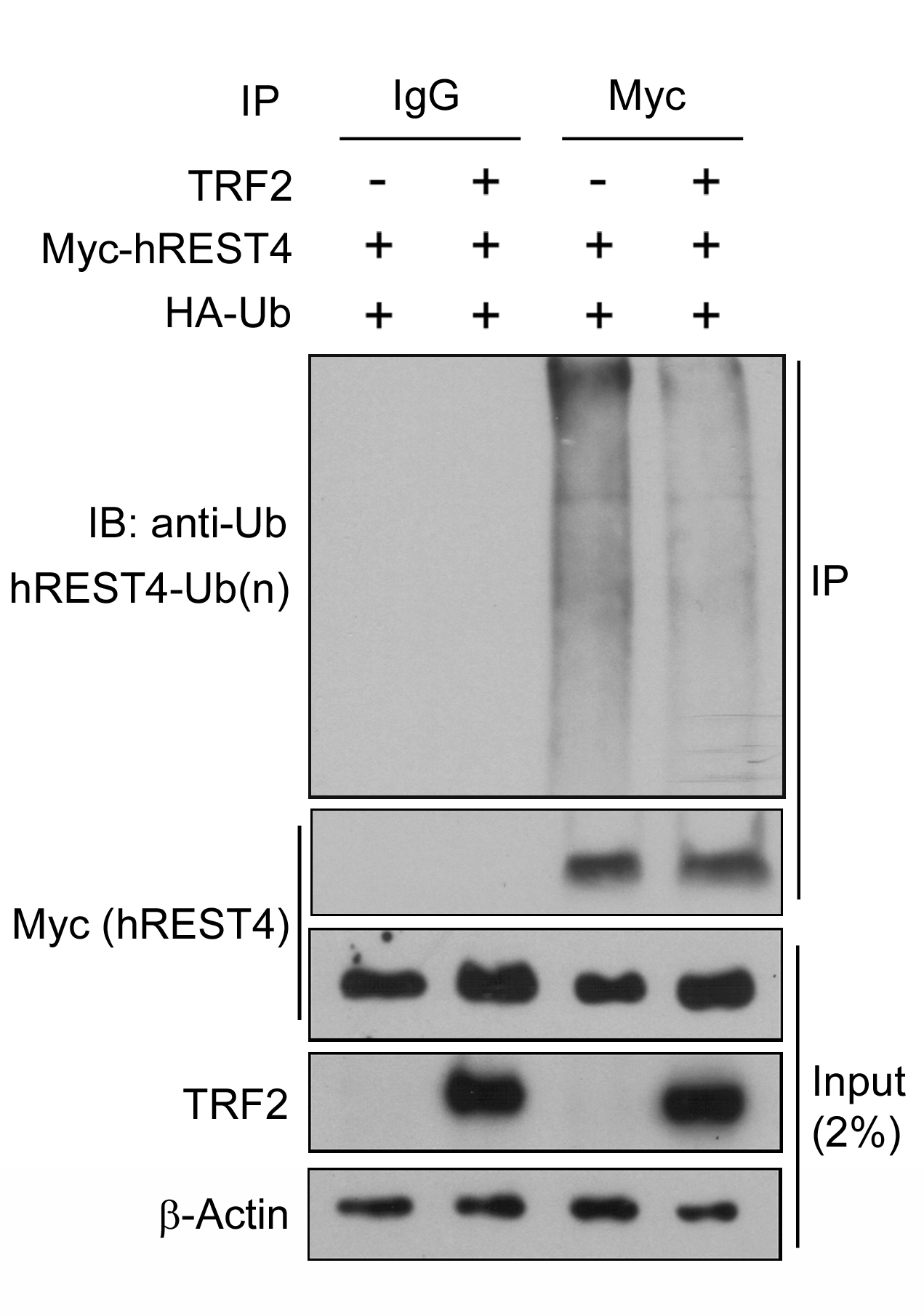
Figure 1. TRF2 inhibits human REST4 ubiquitination. Control and TRF2 stably overexpressing HEK293T cells were co-transfected with Myc-REST4 and HA-ubiquitin (HA-Ub) expression vectors. Myc immunoprecipitates (IP) or whole cell lysates (input) were immunobloted with the indicated antibodies. Immunoblot is representative of two independent experiments.
Recipes
- Antibody binding buffer
PBS + 0.01% Tween 20 (1 µl Tween 20 to 10 ml PBS) - HEPES lysis buffer
20 mM HEPES (pH 7.4)
10 mM KCl
100 mM NaCl2
1 mM MgCl2
1 mM EDTA
0.1 mM EGTA
0.5 mM CaCl2
Before use add following reagents to the indicated final concentrations: 1x protease inhibitor cocktail, 0.2 mM phenylmethanesulfonyl fluoride, 0.1% Triton X-100 and 0.5% Nonidet P40 - 2x electrophoresis sample buffer
2.5 ml of 0.5 M Tris-HCl (pH 6.8)
2 ml of glycerol
4 ml of 10% SDS (w/v)
0.94 ml dH2O
0.16 ml 1.25 % bromophenol blue
Total volume: 9.5 ml
To prepare 1 ml, add 50 µl of -mercaptoethanol to 950 µl 2x sample buffer
Acknowledgments
We are grateful to Dr. Shideng Bao from Lerner Research Institute for the REST plasmid. The work was supported by the funding of BBSRC, MRC (G0802537) and Genesis Research Trust. This work is adapted from the work previously published (Ovando-Roche et al., 2014).
References
- Bhattacharyya, S., Yu, H., Mim, C. and Matouschek, A. (2014). Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol 15(2): 122-133.
- Huang, Z., Wu, Q., Guryanova, O. A., Cheng, L., Shou, W., Rich, J. N. and Bao, S. (2011). Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 13:142-152.
- Ovando-Roche, P., Yu, J. S., Testori, S., Ho, C. and Cui, W. (2014). TRF2-mediated stabilization of hREST4 is critical for the differentiation and maintenance of neural progenitors. Stem Cells 32(8): 2111-2122.
- Ristic, G., Tsou, W. L. and Todi, S. V. (2014). An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes. Front Mol Neurosci 7: 72.
- Weissman, A. M., Shabek, N. and Ciechanover, A. (2011). The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol 12(9): 605-620.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cui, W. and Ovando-Roche, P. (2015). Identification of Factors in Regulating a Protein Ubiquitination by Immunoprecipitation: a Case Study of TRF2 on Human REST4 Ubiquitination. Bio-protocol 5(13): e1525. DOI: 10.21769/BioProtoc.1525.
Category
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









