- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Immunoprecipitation Using Nicotiana benthamiana Transient Expression System
Published: Vol 5, Iss 13, Jul 5, 2015 DOI: 10.21769/BioProtoc.1520 Views: 23193
Reviewed by: Fanglian HeCindy Ast

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Chemiluminescence Based Receptor-ligand Binding Assay Using Peptide Ligands with an Acridinium Ester Label
Mari Wildhagen [...] Markus Albert
Mar 20, 2015 11680 Views

Mating Based Split-ubiquitin Assay for Detection of Protein Interactions
Wijitra Horaruang and Ben Zhang
May 5, 2017 17315 Views

Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
Anna Karlsson [...] Elton P. Hudson
May 5, 2025 2011 Views
Abstract
Nicotiana benthamiana (N. benthamiana) is a useful model system to transiently express protein at high level. This protocol describes in detail how to transiently express protein in N. benthamiana and how to carry out protein immunoprecipitation in this expression system. This protocol can be broadly used for investigation on protein-protein interaction, protein purification and other related protein assay.
Materials and Reagents
- Nicotiana benthamiana seeds
- Agrobacterium strains harboring binary vector (e.g. pCambia1300, pBIN19) with the targeted gene fused with protein purification tag
Note: In this protocol, we use 3x FLAG tag as an example. - Acetosyringone (Sigma-Aldrich, catalog number: D134406 )
- MES (pH 5.6) (Sigma-Aldrich, catalog number: M8250 )
- Protease Inhibitor (Roche, catalog number: 04693132001 )
- Nonidet P 40 substitute (Fluka, catalog number: 74385 )
- Induction medium (see Recipes)
- Infiltration solution (see Recipes)
- Extraction buffer (see Recipes)
- TBS (see Recipes)
- 4x SDS (see Recipes)
Equipment
- Anti-FLAG beads (Sigma-Aldrich, catalog number: A2220 )
- 3x FLAG peptides (Sigma-Aldrich, catalog number: F3290 )
- Protein G sepharose beads (GE healthcare, catalog number: 17-0618-01 )
- Test tubes, flask and shaker machine for culturing Agrobacterium
- Centrifuge
- 1 ml blunt syringe (BD, catalog number: 309659 )
Procedure
- Grow Nicotiana benthamiana at 22 °C (16 h day/ 8h night) to the age of 4 weeks. Grow Agrobacterium strains harboring binary vector with the targeted gene fused with 3x FLAG overnight at 28 °C at 200 rpm in 3 ml LB with proper antibiotic selection. With a sterilized pipette tip, inoculate a tiny amount of Agrobacterium glycerol stock into the test tube.
- Prepare induction medium and inoculate agrobacterium from the LB medium into the induction medium. (1/100 dilution for overnight culture or 1/25 for about 8 h culture). This can be done in the morning to prepare for an evening infiltration, or at night to prepare for infiltration the next morning.
- Incubate at 28 °C, with shaking at 200 rpm, for ~8-12 h, so that the Agrobacterium cell is in the log growth phase. The best OD value should be 0.6-0.8.
- Centrifuge Agrobacterium culture in 50 ml Falcon tube for 10 min at 3,200 x g to pellet the cells.
- Prepare infiltration solution.
- Resuspend Agrobacterium pellet in ~5-10 ml of infiltration solution.
- Measure the OD of Agrobacterium cell and add appropriate amounts of infiltration solution to dilute the Agrobacteria to the desired OD (usually 0.1-0.3 OD). Make at least 1 ml infiltration solution per leaf. If you want to co-express two or multiple proteins, mix different kinds of Agrobacteria, with the desired OD for each one. It’s recommended to carry out preliminary expression experiment to test the protein expression level for different constructs. You can adjust the OD value for each kind of Agrobacteria based on its protein expression level in N. benthamiana. For instance, you can use larger OD value for Agrobacteria expressing lower protein while use smaller OD value for Agrobacteria expressing abundant protein.
- Water the N. benthamiana plants about 30 min before the infiltration to make the infiltration easier.
- Choose the proper leaves for infiltration. Choose the healthiest looking plants, and avoid leaves that are torn or otherwise damaged. The ideal leaves are the young leaves, ~3-5 cm across, use 2-3 leaves per plant. One leaf is about 0.1 g of tissue, which is enough for measuring protein expression. For immunoprecipitation, 10 leaves are usually enough.
- Use a 1 ml blunt syringe to inject the solution into the underside of the leaf. Swirl the solution in the tube before drawing it up into the syringe, in order to evenly suspend the agrobacterium. Try to be as gentle as possible when injecting, to avoid damaging the leaf. The infiltrated area can easily be seen as a dark, water-soaked region. If the infiltrated region stops expanding without pushing too hard, infiltrate again in a different area of the leaf. There is no specific requirement for the infiltration regions as long as the infiltration can cover the leaf. Try to finish the infiltration using less than 5 different infiltrations per leaf. There is short video showing the Agrobacteria infiltration of N. benthamiana: https://www.youtube.com/watch?v=GHc7PU_jG2M. After infiltration, the plants can be grown at the same condition as before.
- Harvest the N. benthamiana leaves without the main vein at 36-48 h after agrobacterium infection. 1 g tissue usually is enough. Time point experiment might need to be carried out to decide at which time point the protein express the highest.
- Grind the tissues into a fine powder in liquid nitrogen using a cold mortar and pestle.
- Add extraction buffer [see Recipes, with 2% PVPP (polyvinylpolypyrolidone), 10 mM DTT, 1x protease inhibitor (PI) and 1 mM PMSF] into the mortar (2 ml extraction buffer per gram plant tissue), place it in the 4 °C fridge, wait until the powder start to thaw and homogenize the mixture by grinding.
- Transfer all the liquid into the 1.5 tubes and centrifuge at highest speed for 10 min at 4 °C.
- Transfer the supernatant into new tubes and centrifuge for another time.
- Transfer the supernatant into the tubes and add 20% NP40 to a final concentration of 0.15%.
- Add 20 μl protein G sepharose beads into the sample and incubate for 30 min in the cold room to pre-clear the sample (the beads were pre-equilibrated using 1 ml extraction buffer and the supernatant buffer was discarded after centrifugation at 12,000 x g for 30 sec).
- Pellet the beads by centrifugation at 12,000 x g for 30 sec and transfer the supernatant into new tubes. 60 μl of the supernatant is taken as the input sample, which stands for the sample before immunoprecipitation.
- Add 10 μl anti-FLAG beads into the supernatant. The beads also need to be equilibrated as shown in step 18. Incubate the beads at 4 °C for 3 h on an Eppendorf tube rotator.
- Pellet the beads by centrifugation and take 60ul of supernatant for flow-through sample.
- Wash the beads with 1 ml extraction buffer (with 0.15% NP40) for 3 times. For each washing step, pellet the beads by centrifuging at 12,000 x g for 1 min at 4 °C, discard the supernatant and add 1 ml extraction buffer.
- Add 100 μl diluted 3x FLAG peptide (dilute the 3x FLAG stock 1/10 in TBS containing 1 mM EDTA with 1x protease inhibitor. The 3x FLAG stock is 2.5 mg/ml in TBS which can stored at -20 °C for years) into the beads and incubate in the cold room for 1 h on an Eppendorf tube rotator.
- Pellet the beads by centrifugation. Transfer the supernatant into a new tube. Add 35 μl 4x SDS loading buffer into the supernatant. This fraction is the elution sample containing the purified immunoprecipitated protein. Add 60 μl 1x SDS loading buffer into the tube containing the beads. The beads sample might contain the residue protein that fails to elute from the beads.
- Add proper volume of 4x SDS loading buffer to the samples of input and flow-through. Boil all the samples at 95-100 °C for 5-10 min. And analyze the samples using Western blot.
Representative data
A representative experiment result was shown in the Figure 1. The figure was adapted from the Xu et al. (2014) as listed in the reference.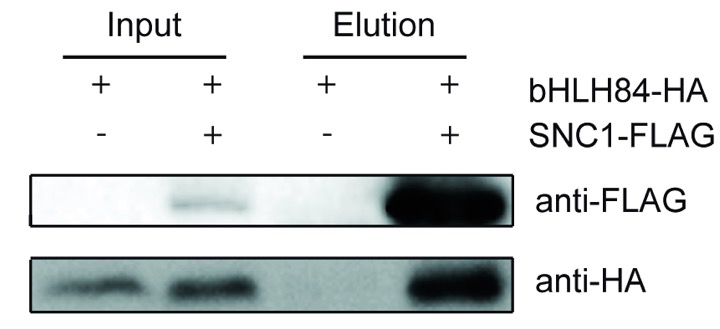
Figure 1. bHLH84-HA co-immunoprecipitates with SNC1-FLAG. Two samples were used in the Co-IP experiment including sample with coexpression of bHLH84 and SNC1-FLAG and sample with expression of only bHLH84, which serves as the negative control.
Notes
- Negative control samples are always recommended during the immunoprecipitation process. For immunoprecipitation of single protein, it is suggested to express the empty vector to serve as the negative control. For co-immunoprecipitation of two or more proteins, it is suggested to express the empty vector without the bait protein together with the relative vectors containing the prey proteins. Based on the experiment design, more negative controls can be included accordingly.
- As long as the protein can be pulled down, the immunoprecipitation experiment is highly reproducible although the IP efficiency might be vary slightly in different trials.
Recipes
- Induction medium
Note: You also need to add proper antibiotics to the following media.
Reagent Stock concentration Sterilization Volume /100 ml Final concentration K2HPO4 10.5 g/L Mix all together with water and autoclave 96.85 ml KH2PO4 4.5 g/L (NH4)2SO4 1.0 g/L NaCitrate:2H2O 0.5 g/L Glycerol 0.5% Glucose 20% Filter 1 ml 0.2% MgSO4 1 M Filter 100 µl 1 mM Acetosyringone 100 mM Filter 50 µl 50 µl MES (pH 5.6) 0.5 M Filter 2 ml 10 mM - Infiltration solution
Reagent Stock concentration Sterilization Amount per 20 ml Final concentration MS powder NA 0.088 g 4.4 mg/L MES 0.5 M Filter 0.4 ml 10 mM Acetosyringone 100 mM Filter 30 µl 150 µM - Extraction buffer
10% glycerol
25 mM Tris-HCl (PH 7.5)
1 mM EDTA
150 mM NaCl - TBS
10 mM Tris-HCl
150 mM NaCl (PH 7.4) - 4x SDS
200 mM Tris-HCl (PH 6.8)
8% SDS
40% glycerol
0.04% bromophenol blue
400 mM DTT
Acknowledgments
We are grateful for the financial supports from Natural Sciences and Engineering Research Council (NSERC) (Canada) Discovery grant program. The transient protein expression in N. benthamiana was adapted from Van den Ackerveken et al. (1996). The IP protocol was modified from Moffett et al. (2002).
References
- Moffett, P., Farnham, G., Peart, J. and Baulcombe, D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21(17): 4511-4519.
- Van den Ackerveken, G., Marois, E. and Bonas, U. (1996). Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87(7): 1307-1316.
- Xu, F., Kapos, P., Cheng, Y. T., Li, M., Zhang, Y. and Li, X. (2014). NLR-associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PLoS Pathog 10(8): e1004312.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Xu, F., Copeland, C. and Li, X. (2015). Protein Immunoprecipitation Using Nicotiana benthamiana Transient Expression System. Bio-protocol 5(13): e1520. DOI: 10.21769/BioProtoc.1520.
Category
Plant Science > Plant biochemistry > Protein > Interaction
Biochemistry > Protein > Immunodetection > Immunoprecipitation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









