- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Murine Liver Myeloid Cell Isolation Protocol
(*contributed equally to this work) Published: Vol 5, Iss 10, May 20, 2015 DOI: 10.21769/BioProtoc.1471 Views: 33115
Reviewed by: Fanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2243 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views
Abstract
In homeostasis, the liver is critical for the metabolism of nutrients including sugars, lipids, proteins and iron, for the clearance of toxins, and to induce immune tolerance to gut-derived antigens. These functions predispose the liver to infection by blood-borne pathogens, and to a variety of diseases ranging from toxin and medication-induced disorders (CCl4, acetaminophen) to metabolic disorders (steatohepatitis, alcoholic liver disease, biliary obstruction, cholestasis) or autoimmunity. Chronic liver injury often progresses to life threatening fibrosis and can end in liver cirrhosis and hepatocellular carcinoma (Pellicoro et al., 2014).
The liver contains parenchymal cells or hepatocytes that make up the majority of hepatic cells. It also contains non-parenchymal structural cells such as sinusoidal endothelial cells and a large number of non-parenchymal innate immune cells, mainly monocytes, neutrophils, macrophages, DCs, NK and NKT cells that can trigger an adaptive immune response in the case of infections or other pathogenic insults (Jenne and Kubes, 2013). How this immune response is regulated determines the extent of acute and chronic liver injury (Stijlemans et al., 2014). In this context, liver macrophages have been demonstrated to play central but divergent (from initiating to resolving) functions in liver injury (Sica et al., 2014). It has become clear in the last years that hepatic macrophages consist of two classes, tissue-resident macrophages, the Kupffer cells (KCs) originating from yolk sac/fetal liver progenitors and tissue-infiltrating macrophages originating from bone marrow-derived Ly6CHi monocytes (Jinhoux and Jung, 2014; Tacke and Zimmerman, 2014). Distinguishing the activities of KCs from those of monocyte-derived macrophages during liver injury or repair is currently a frontline research topic in the macrophage field. Indeed, considering that clinical management of liver failure remains problematic, a better understanding of the immune mechanisms regulating liver injury is expected to allow the development of new therapeutic modalities. Here, we describe an isolation technique for liver non-parenchymal polymorphonuclear (PMN) and mononuclear myeloid cells permitting their molecular and functional characterization.
Materials and Reagents
- 7-8 weeks old female C57Black/6 mice (Janvier Labs)
- RPMI-1640 medium (RPMI) (Life Technologies, catalog number: 52400-041 )
- Collagenase Type III (Worthington Biochemical, catalog number: LS004180 )
- DNase I (Roche Diagnostics, catalog number: 04536282001 )
- Heparin (sodium salt from porcine intestinal mucosa) (Sigma-Aldrich, catalog number H3393-1MU )
- Hank’s buffered salt solution (HBSS) without calcium or magnesium or phenol red (Life Technologies, Gibco®, catalog number: 14175-053 )
- NH4Cl (Merck KGaA, catalog number: 01145.0500 )
- KHCO3 (Merck KGaA, catalog number: 04854.0500 )
- EDTA (Duchefa Biochemie, catalog number: E0511.1000 )
- HCl (37% stock solution) (Merck KGaA, catalog number: 1.00317.1000 )
- Fetal bovine serum (FBS) (BiowhittakerTM/Lonza, catalog number: DE14-801F )
- LymphoprepTM (Axis-shield, catalog number: 1114547 )
- PercollTM (GE Healthcare, catalog number: 17-0891-01 )
- Purified CD16/CD32 (Fc-Block) (clone 2.4G2) (BD Biosciences, catalog number: 553142 )
- PE-Cy7-conjugated anti-CD11b antibody (clone M1/70) (BD Biosciences, catalog number: 552850 )
- AF647-conjugated anti-Ly6C antibody (clone ER-MP20) (Serotec, catalog number: MCA2389A647 )
- PerCP-Cy5.5-conjugated anti-I-A/I-E (MHC-II) antibody (clone M5/114.15.2) (Biolegend, catalog number: 107626 )
- FITC-conjugated anti-Ly6G antibody (clone 1A8) (BD Biosciences, catalog number: 551460 )
- APC-Cy7-conjugated CD45 antibody (clone 30-F11) (BD Biosciences, catalog number: 103116 )
- PE-conjugated F4/80 antibody (clone CI: A3-1) (AbD Serotec, catalog number: MCA497PET )
- Trypan blue (BDH Chemicals, catalog number: 34078 )
- NaCl (Thermo Fisher Scientific, catalog number: 10428420 )
- KH2PO4 (Merck KGaA, catalog number: 1.04873.1000 )
- Na2HPO4.2H2O (Merck KGaA, catalog number: 1.06580.1000 )
- L-glutamine (Sigma-Aldrich, catalog number: G8540-100G )
- Penicillin (Life Technologies, Gibco®, catalog number: 15140-122 )
- Streptomycin (Life Technologies, Gibco®, catalog number: 15140-122)
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- Sodium pyruvate (Life Technologies, Gibco®, catalog number: 11360-039 )
- Non-essential amino acids (Life Technologies, Gibco®, catalog number: 11140-035 )
- Liver digestion medium (see Recipes)
- Phosphate buffered saline (PBS) (see Recipes)
- 33% Percoll working solution (see Recipes)
- Erythrocyte lysis buffer (see Recipes)
- MACS buffer (see Recipes)
- Cell suspension medium (see Recipes)
- Blocking medium (see Recipes)
- Complete medium (see Recipes)
- Trypan blue working solution (see Recipes)
Equipment
- Polyester filters cut in 10 x 10 cm squares, thread diameter 70 μm (Spectrumlabs, catalog number: 146490 )
- 10 ml syringes (Omnifix, catalog number: 473203 )
- BD Falcon 50 ml polypropylene tubes (BD Biosciences, catalog number: 2070 )
- BD Falcon 15 ml polypropylene tubes (BD Biosciences, catalog number: 2096 )
- BD Falcon 5 ml polypropylene round-bottom tube (BD Biosciences, catalog number: 352063 )
- Needles (Microlance 22G1 ½, 0.7 * 40 mm) (BD Biosciences, ref: 301000 )
- Sterile culture hood
- Surgical scissors and forceps
- 37°C, 5% CO2 cell culture incubator (Forma Scientific)
- Pipettes
- Centrifuges (Eppendorf, models: 5810R and 5417C )
- Orbital shaker (Belgolabo, model: Julabo type SW-20C ) used at 200 rpm
- Light microscope (Olympus, model: CK2 )
- Multicolor flow cytometer (BD Biosciences, FACSCantoTM )
- GentleMACSTM Dissociator (Miltenyi Biotec, catalog number: 130-093-235 )
- GentleMACSTM C-tubes (Miltenyi Biotec, catalog number: 130-093-237 )
Procedure
- Preparation of a liver single cell suspension
- Sacrifice the mouse using CO2 and restrain it by pinning its paws into a foam surface using syringe needles. Of note, all murine experiments were performed according to the ECPVA guidelines (CETS n° 123) and were approved by the VUB Ethical Committee (Permit Number: 08-220-8). Heparinized blood was taken via cardiac puncture (~1 ml) in order to prevent too much blood contamination when taking the liver. Alternatively, the liver can be perfused in vivo via the portal vein with 10 ml saline. Make a parallel incision from the base of the tail up to the neck along the mouse’s abdomen and to the paws without puncturing the peritoneum. Gently pull back the skin and pin it to the foam surface. Subsequently, open the mouse abdomen and softly move the intestines on the side to get access to the liver using a cotton plug.
- Gently take out the liver from the body, without damaging it.
- Store the harvested liver in 5 ml RPMI medium in a 50 ml Falcon tube on ice until the digestion procedure.
- Put the liver in a GentleMACSTM C-tube and add 5 ml liver digestion medium. Subsequently, cut the liver in small pieces (1-1.5 mm) using scissors (Figure 1A/B).
- Homogenize the liver using the GentleMACSTM Dissociator (Figure 1C) using 2 times program mLiver_01_03 (16 sec) at room temperature. Subsequently, incubate at 37 °C for 20-30 min while shaking in a water bath to allow digestion of the tissue.
- Homogenize the liver suspension once more using the GentleMACSTM Dissociator program mLiver_02_03 (25 sec) at room temperature (Figure 1D). Finally, add 5 ml of blocking medium to stop the liver digestion.
- In order to measure the cytokine content within whole liver, collect 0.5 ml of the solution in a 1.5 ml Eppendorf tube and centrifuge it at 10,625 x g for 8 min at room temperature. Subsequently, collect the supernatant and store at -20 °C till needed.
- Filter the remaining liver suspension through a 70-µm sterile nylon gauze into a sterile 50 ml conical tube. Wash the GentleMACSTM C-tube with an additional 20 ml of blocking solution and rinse the filter once more with this.
- Centrifuge the 50 ml tubes at 450 x g for 8 min at 4 °C and gently discard the supernatant.
- Eliminate the red blood cells by resuspending the cell pellet in 5 ml ice-cold erythrocyte lysis buffer and leaving it on ice for 2-3 min.
- Neutralize the lysis by adding 25 ml cell suspension medium, and transfer the suspension to a new 50 ml tube through a 70-µm sterile nylon gauze.
- Centrifuge the 50 ml tube at 450 x g for 8 min at 4 °C and gently discard the supernatant.
- Resuspend the cell pellet in 5 ml cell suspension medium and count the living cells using Trypan blue.
At this stage, the suspension contains both parenchymal (hepatocytes) and non-parenchymal (including polymorphonuclear cells, monocytes, macrophages, DCs, NK and NKT cells) liver cells. Although this suspension can be analyzed via flow-cytometry, it is advisable to perform additional fractionation steps to allow cell culturing and/or FACS sorting.
- Sacrifice the mouse using CO2 and restrain it by pinning its paws into a foam surface using syringe needles. Of note, all murine experiments were performed according to the ECPVA guidelines (CETS n° 123) and were approved by the VUB Ethical Committee (Permit Number: 08-220-8). Heparinized blood was taken via cardiac puncture (~1 ml) in order to prevent too much blood contamination when taking the liver. Alternatively, the liver can be perfused in vivo via the portal vein with 10 ml saline. Make a parallel incision from the base of the tail up to the neck along the mouse’s abdomen and to the paws without puncturing the peritoneum. Gently pull back the skin and pin it to the foam surface. Subsequently, open the mouse abdomen and softly move the intestines on the side to get access to the liver using a cotton plug.
- Fractionation of liver cells using a Lymphoprep gradient separation
- Add slowly 10 ml LymphoprepTM (Lucron Bioproducts) underneath the resuspended liver cell solution using a 10 ml syringe with long needle. Centrifuge at 800 x g for 25 min at 20 °C without acceleration or brake (Figure 1E).
- Carefully collect the layer of low-density cells at the interphase containing the non-parenchymal liver cells (enriched in mononuclear myeloid cells as well as T-, B- and NK/NKT-cells). If the interphase is not clearly visible (low amount of cells) you can also collect the upper phase (containing RPMI medium). Take as less as possible of the lower phase containing Lymphoprep (Figure 1E).
- Transfer the interphase to a new sterile 15 ml tube and fill to the top with MACS buffer. Centrifuge at 800 x g for 7 min at 20 °C and discard the supernatant. Resuspend the cell pellet in 1-2 ml cell suspension medium, and after counting the cells using Trypan blue, bring at a concentration of 107 cells/ml.
- After removal of the Lymphoprep, collect the lower fraction (pellet) containing mainly parenchymal cells (hepatocytes) and polymorphonuclear (PMN) cells that were not retained within the non-parenchymal cell fraction due to their high density characteristics and transfer it to a sterile 15 ml Falcon tube (Figure 1E). Resuspend the cells in a final volume of 15 ml cell suspension medium and centrifuge at 650 x g for 8 min at 20 °C.
- Subsequently, in order to remove the Lymphoprep solution perform 1-2 washing steps of 15 ml cell suspension medium. Finally, resuspend the pellet in 3 ml cell suspension medium and, after counting the cells using Trypan blue, bring at a concentration of 107 cells/ml.
The interphase (non-parenchymal cells) fraction can now be used for FACS analysis/sorting as well as for cell culturing conditions. The bottom fraction containing hepatocytes and remaining non-parenchymal cells (PMN as well as macrophages with higher density) can also be used for flow-cytometric analysis. The latter fraction can be further cleaned-up using a Percoll gradient in order to separate hepatocytes from the remaining non-parenchymal cells. Of note, if hepatocytes are not needed DNase 1 can be added to the digestion medium, resulting in a cleaner sample.
- Add slowly 10 ml LymphoprepTM (Lucron Bioproducts) underneath the resuspended liver cell solution using a 10 ml syringe with long needle. Centrifuge at 800 x g for 25 min at 20 °C without acceleration or brake (Figure 1E).
- Fractionation of liver cells using a Percoll gradient separation
- To separate hepatocytes from the PMN and remaining macrophages, mix 1 vol. (~ 5 ml of the 107/ml working solution) of cell suspension with 1 vol. of isotonic Percoll solution of density 1.07 g/ml (33% working solution). Centrifuge for 30 min at 800 x g at room temperature (Figure 1F). Aspirate hepatocytes, forming a broad band at a density of 1.07-1.09 g/ml, from the gradient and wash free of Percoll by two additional cycles of centrifugation at 800 x g, for 5 min and resuspension in 3 ml cell suspension medium.
- After removal of the Percoll, collect the lower cell pellet fraction containing mainly PMN and macrophages that were not retained within the non-parenchymal fraction due to their density characteristics and transfer it to a sterile 15 ml Falcon tube.
- Subsequently, centrifuge at 650 x g for 5 min. at 4 °C, resuspend the pellet in 1 ml cell suspension medium, count the cells using Trypan blue and bring them at a concentration of 107 cells/ml.
The different fractions obtained in sections B and C can be used for flow-cytometric analysis or when resuspended in complete medium for in vitro cell culturing.
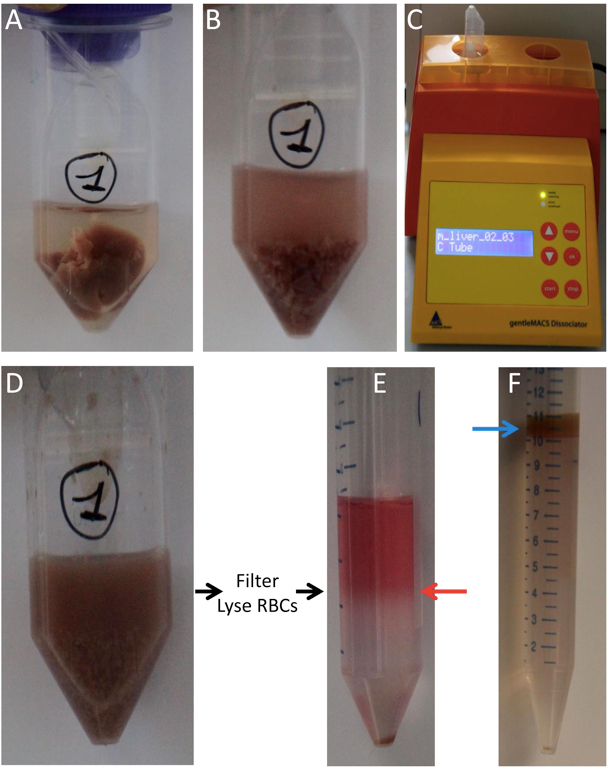
Figure 1. Liver single-cell preparation. A. Liver in a GentleMACSTM C-tube in 5 ml digestion medium. B. Liver in a GentleMACSTM C-tube after cutting with scissors. C. GentleMACSTM Dissociator. D. Liver in a GentleMACSTM C-tube after digestion and homogenization using the GentleMACSTM Dissociator. Subsequently, filter the suspension and lyse RBCs. E. Gradient after centrifugation with a clearly visible interphase (non-parenchymal cells, red arrow) and a pellet (hepatocytes and remaining non-parenchymal cells with higher density). F. Optional: Percoll gradient after centrifugation with a clearly visible upper fraction (hepatocytes, blue arrow) and a pellet (PMN and remaining non-parenchymal cells with higher density).
- To separate hepatocytes from the PMN and remaining macrophages, mix 1 vol. (~ 5 ml of the 107/ml working solution) of cell suspension with 1 vol. of isotonic Percoll solution of density 1.07 g/ml (33% working solution). Centrifuge for 30 min at 800 x g at room temperature (Figure 1F). Aspirate hepatocytes, forming a broad band at a density of 1.07-1.09 g/ml, from the gradient and wash free of Percoll by two additional cycles of centrifugation at 800 x g, for 5 min and resuspension in 3 ml cell suspension medium.
- Flow-cytometric analysis
- Transfer 100 µl of a 107 cells/ml stock solution of the different fractions [(interface Lymphoprep (section B), pellet Percoll (section C)] into a 5 ml polypropylene round-bottom tube to prevent sticking of cells. Incubate the cell suspension with 1 µg rat anti-mouse CD16/CD32 FcR-blocking antibody clone (2.4 G2, 1 µg per 106 cells) on ice water for 20 min.
- Subsequently, add fluorescently labeled antibodies (0.2 µg per 106 cells) for another 20 min on ice water, protected from exposure to light. Antibodies used are FITC-conjugated Ly6G, PE-conjugated F4/80, APC-conjugated Ly6C, APC-Cy7 conjugated CD45, PE-Cy7-conjugated anti-CD11b and PerCP-Cy5.5-conjugated MHC-II.
- Wash by adding 2 ml ice-cold MACS buffer, centrifuge at 450 x g for 6 min at 4 °C and discard the supernatant. Add 100-200 µl cell suspension medium to keep cells alive, transfer the cells into a FACS tube and proceed to the FACSCantoTM.
- Analyze the FACS data using FlowJo software. Briefly, after selecting a life gate and single cells (using a FSC-A versus FSC-H profile) select the CD45+ cells in a CD45 versus FSC-A plot (Figure 2, upper and lower panels). Within the CD45+ cells, gate out the PMN based on their CD11bpos Ly6Gpos expression profile and select for the CD11bpos Ly6Gneg expressing cells (Figure 3A). The CD11bneg Ly6Gneg population consists mainly of T-cells, B-cells and NK-cells. Next, plot the CD11bpos Ly6Gneg cells in a Ly6C versus MHC-II plot and subsequently check for F4/80 expression. As such, monocytes as Ly6Chigh MHC-IIneg F4/80low cells, monocyte-derived “immature” macrophages as Ly6Chigh MHC-IIhigh F4/80high cells, resident/mature macrophages (i.e. Kupffer cells/”mature” monocyte-derived macrophage) as Ly6Cneg/low MHC-IIhigh F4/80high cells, patrolling monocytes as Ly6Cneg/low MHC-IIneg F4/80low cells and eosinophils based on their Ly6Cint MHC-IIneg F4/80low expression are identified (Figure 3B). Of note, the identity of eosinophils is confirmed based on their SiglecF expression and high SSC.
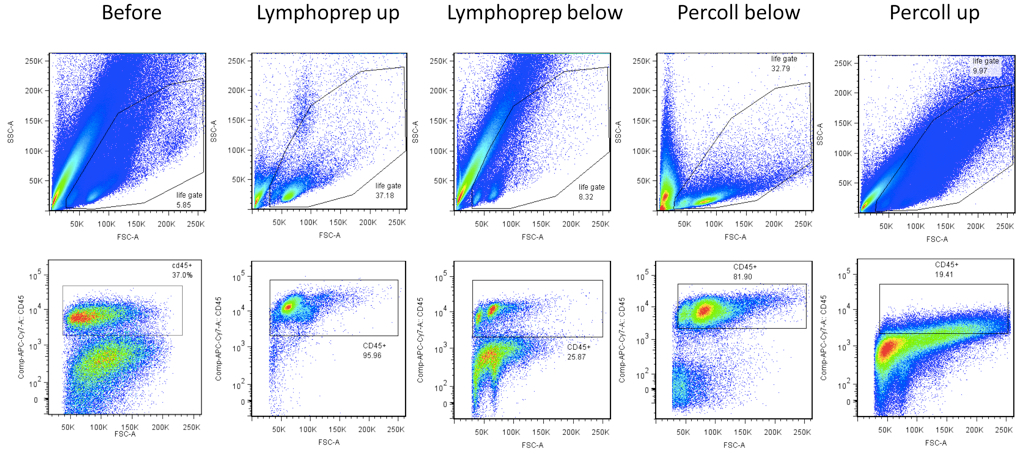
Figure 2. Representative FACS gating strategy for different liver preparations (total liver suspension, Lymphoprep upper fraction, Lymphoprep lower fraction, Percoll lower fraction and Percoll upper fraction). (Upper) Selection of a life gate based on a SSC-A versus FSC-A plot. (Lower) After selection of single cells, select CD45+ cells in a CD45 versus FSC plot.
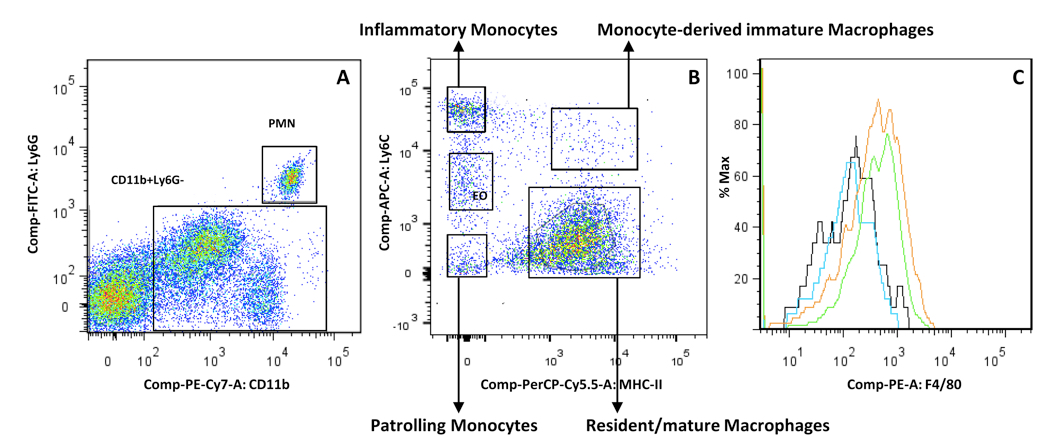
Figure 3. Representative gating strategy for myeloid cells within the CD45+ fraction. A. Plot the CD45+ cells (see Figure 2, lower panels) in a Ly6G versus CD11b plot to identify PMN (CD11bpos Ly6Gpos), mononuclear cells (CD11bpos Ly6Gneg) and CD11bneg Ly6Gneg cells (T-cells, NK-cells, B-cells). B. The remaining CD11bpos Ly6Gneg fraction is plotted in a MHC-II versus Ly6C plot to allow identifying monocytes (Ly6ChighMHC-IIneg), monocyte-derived “immature” macrophages (Ly6Chigh MHC-IIhigh), “resident/mature” macrophages (Kupffer cells, Ly6Cneg/low MHC-IIhigh), patrolling monocytes (Ly6Clow/negMHC-IIneg) and eosinophils (EO, Ly6CintMHC-IIneg). C. F4/80 plot of the inflammatory monocytes (black), patrolling monocytes (blue), Monocyte-derived “immature” macrophages (green) and “resident/mature” macrophages (orange).
- Transfer 100 µl of a 107 cells/ml stock solution of the different fractions [(interface Lymphoprep (section B), pellet Percoll (section C)] into a 5 ml polypropylene round-bottom tube to prevent sticking of cells. Incubate the cell suspension with 1 µg rat anti-mouse CD16/CD32 FcR-blocking antibody clone (2.4 G2, 1 µg per 106 cells) on ice water for 20 min.
Recipes
- Liver digestion medium
1 ml Collagenase Type III stock: 6,000U/ml) diluted in 50 ml Hanks' Balanced Salt Solution (HBSS) without calcium or magnesium
Aliquot the stock solution of Collagenase Type III (1 ml/tube) and freeze at -20 °C
Optional: If hepatocytes are not needed add 10 Units/ml of DNase I to the medium.
- Phosphate buffered saline (PBS)
8 g/L NaCl
0.2 g/L KCl
0.24 g/L KH2PO4
1.8 g/L Na2HPO4.2H2O
Add distilled water till 1 L and adjust pH: 7.4 with HCl (stock solution 37%)
- 33% Percoll working solution
Add PBS to 16.5 ml PercollTM stock solution till 50 ml final volume
- Erythrocyte lysis buffer
8.29 g/L NH4Cl
1 g/L KHCO3
37.2 mg/L EDTA
Add distilled water till 1 l and bring at pH 7.2 using HCl
- MACS buffer
HBSS without calcium or magnesium or phenol red
2% (v/v) heat-inactivated fetal bovine serum (FBS)
3 mM EDTA
- Cell suspension medium
RPMI
5% (v/v) heat-inactivated fetal bovine serum (FBS)
- Blocking medium
HBSS without calcium or magnesium or phenol red
2% (v/v) heat-inactivated FBS
5 mM EDTA
- Complete medium
Roswell Park Memorial Institute (RPMI)-1640
10% (v/v) heat-inactivated fetal bovine serum (FBS)
300 μg/ml L-glutamine
100 U/ml penicillin
100 μg/ml streptomycin
0.02 mM β-mercaptoethanol
1 mM sodium pyruvate
1 mM non-essential amino acids
- Trypan blue working solution
Make a 1% v/v Trypan blue solution in PBS and use it in a 1/10 ratio (Cells/Trypan blue)
Acknowledgments
We acknowledge the financial support of the Interuniversity Attraction Pole Program (PAI-IAP N. P7/41, http://www.belspo.be/belspo/iap/index_en.stm) and a grant from the FWO (KaN 1511812N). Benoit Stijlemans is a research fellow supported by the VUB/SRP Targeting inflammation linked to infectious diseases and cancer (Nanobodies for Health). The authors also thank Ella Omasta, Marie-Therese Detobel, Maria Slazak, Victor Orimoloye and Nadia Abou for technical assistance. We also would like to thank Dr. Carl De Trez for his constructive discussions.
References
- Ginhoux, F. and Jung, S. (2014). Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 14(6): 392-404.
- Jenne, C. N. and Kubes, P. (2013). Immune surveillance by the liver. Nat Immunol 14(10): 996-1006.
- Pellicoro, A., Ramachandran, P., Iredale, J. P. and Fallowfield, J. A. (2014). Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol 14(3): 181-194.
- Sica, A., Invernizzi, P. and Mantovani, A. (2014). Macrophage plasticity and polarization in liver homeostasis and pathology. Hepatology 59(5): 2034-2042.
- Stijlemans, B., Leng, L., Brys, L., Sparkes, A., Vansintjan, L., Caljon, G., Raes, G., Van Den Abbeele, J., Van Ginderachter, J. A., Beschin, A., Bucala, R. and De Baetselier, P. (2014). MIF contributes to Trypanosoma brucei associated immunopathogenicity development. PLoS Pathog 10(9): e1004414.
- Tacke, F. and Zimmermann, H. W. (2014). Macrophage heterogeneity in liver injury and fibrosis. J Hepatol 60(5): 1090-1096.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Stijlemans, B., Sparkes, A., Abels, C., Keirsse, J., Brys, L., Elkrim, Y., Baetselier, P. D., Beschin, A. and Ginderachter, J. A. V. (2015). Murine Liver Myeloid Cell Isolation Protocol . Bio-protocol 5(10): e1471. DOI: 10.21769/BioProtoc.1471.
Category
Immunology > Immune cell isolation > Myeloid cell
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link