- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
ELISA Detection of Endogenous Serum Albumin in the Mouse Brain: A Measure of Extravasation Following Brain Injury
Published: Vol 5, Iss 9, May 5, 2015 DOI: 10.21769/BioProtoc.1469 Views: 11440
Reviewed by: Kae-Jiun ChangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Generation of a Human Conditionally Immortalized Cell-based Multicellular Spheroidal Blood-Brain Barrier Model for Permeability Evaluation of Macromolecules
Ryuto Isogai [...] Tomomi Furihata
Aug 5, 2022 3640 Views

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2636 Views
Abstract
After stroke and brain contusion, serum proteins extravasate into nerve tissue through disrupted blood-brain barrier (BBB). Because extravasations of serum proteins result in vasogenic brain edema, serum albumin level in the brain is an indicator of BBB disruption and brain edema after brain insults. In this protocol, extravasation of endogenous albumin is measured in the damaged mouse brain, which would be valuable in the evaluation of vasogenic brain edema formation (Michinaga et al., 2014).
Keywords: Blood-brain barrierMaterials and Reagents
- Mouse Albumin ELISA Quantitation Set (Bethyl Laboratories, catalog number: E90-134 )
Note: This set includes an anti-mouse albumin goat antibody for plate coating, a mouse reference serum and a HRP-conjugated anti-mouse albumin goat antibody. - Mouse serum albumin (Sigma-Aldrich, catalog number: A3139 )
- SureBlueTM TMB microwell peroxidase substrate (Kirkegaard & Perry Laboratories, catalog number: 52-00-01 )
- 450 nm BioFX® liquid Nova-Stop solution for TMB microwell substrates (SurModics, catalog number: LSTP-0100-01 )
- BCA protein assay reagent A (Thermo Fisher Scientific, catalog number: 23221 )
- BCA protein assay reagent B (Thermo Fisher Scientific, catalog number: 23224 )
- Bovine serum albumin (BSA) set (Thermo Fisher Scientific, catalog number: 23208)
- Pentobarbital sodium salt (Nacalai Tesque, catalog number: 26427-14 )
- PBS (Santa Cruz Biotechnology, catalog number: sc-24946 )
- Triton X-100 (Nacalai Tesque, catalog number: 35501-15 )
- Tris (hydroxymethyl) aminomethane (Nacalai Tesque, catalog number: 35409-45 )
- HCl (Nacalai Tesque, catalog number: 18321-05 )
- NaCl (Nacalai Tesque, catalog number: 31320-05 )
- NP-40 (Nacalai Tesque, catalog number: 23640-94 )
- Deoxycholic acid (Wako Chemicals USA, catalog number: 044-18812 )
- SDS (Nacalai Tesque, catalog number: 31607-65 )
- EDTA (Nacalai Tesque, catalog number: 15114-15 )
- Phenylmethylsulfonyl fluoride (Enzo Life Science, catalog number: ALX-270-184-G025 )
- Aprotinin (Sigma-Aldrich, catalog number: A6103 )
- PBS (see Recipes)
- PBST (see Recipes)
- Lysis buffer (see Recipes)
Equipment
- Scissors (Fine Science Tools, catalog number: 14002-14 )
- Forceps (Fine Science Tools, catalog number: 11271-30 )
- Hemostats (Fine Science Tools, catalog number: 13002-10 )
- Winged needle (Terumo, catalog number: SV-27DL )
- Syringe (Terumo, catalog number: SS-30ESZ )
- Razor (FEATHER Safety Razor, catalog number: FH-10 )
- Acrylic brain slicer (Muromachi Kikai, catalog number: MK-MC-01 )
- 1.5 ml sampling tubes (Bio-Bik, catalog number: RC-0170 )
- 15 ml conical tube (BD Biosciences, Falcon®, catalog number: REF 352196 )
- 96-well plate (Thermo Fisher Scientific, catalog number: 167008 )
- Pipette tips (2 to 200 µl) (Eppendorf, catalog number: 0030 073. 800 )
- Stereotactic device (Narishige, model: SR-6N )
- ELISA plate set (Sumitomo Bakelite, catalog number: BS-X7310 )
Note: This set includes ELISA plates, plate seals, an antibody-immobilizing solution and a soaking solution. - Micropipettes (2 to 20 µl and 20 to 200 µl, Eppendorf)
- Microplate reader (Bio-Rad Laboratories, model: 680 )
- Digital Sonifier® (Branson, model: 250D )
- Centrifuge (Hitachi, model: CF15RXII )
- Microplate mixer (AS ONE Corporation, model: NS-4P )
Procedure
- Disruption of BBB by cerebral cold injury [see Material and Methods in Michinaga et al. (2014)]
Mice (5 to 6 week old male ddY, SLC) were placed in a stereotactic device (Narishige, model: SR-6N) under anesthesia. The scalp was incised at the midline and the skull was exposed. A copper rod (10 cm length, 4 mm diameter) was kept in a 50 ml conical tube filled with powdered dry ice. The cooled rod was set on a micromanipulator with the conical tube. The tip of the cooled rod was applied to the skull of the left hemisphere at a position of 3 mm posterior and 5 mm lateral to bregma for 60 sec. In this study, the injured mice were recovered for 72 h prior to perfusion.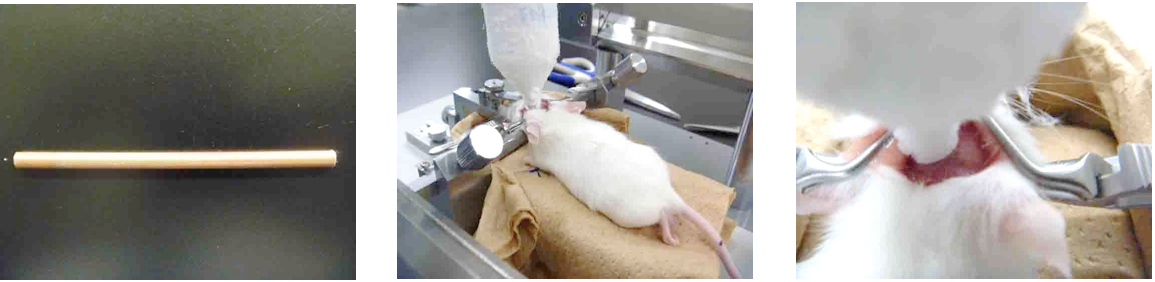
Figure 1. Procedure of cerebral cold injury. A copper rod (10 cm length, 4 mm diameter) was shown in the left photograph. It was set in a 50 ml conical tube and cooled by dry ice powder. After mice were placed in a stereotactic device under anesthesia, the cooled rod was attached in the left hemisphere on the skull shown in middle and right photographs. - Mouse perfusion
- Make experimental manipulations to reproduce brain insults. Disruption of BBB and brain edema will gradually develop thereafter.
- Anesthetize mice deeply by pentobarbital (50 mg/kg, i. p.).
- Perform laparotomy until the heart can be clearly seen.
- Tear the liver for outflow of the circulating blood.
- Connect the wing needle and syringe.
- Prick the winged needle into left cardiac ventricle and slowly inflow 50 ml PBS for about 20 min checking the outflow of blood from the liver.
- Make experimental manipulations to reproduce brain insults. Disruption of BBB and brain edema will gradually develop thereafter.
- Protein samples from the mouse cerebrum
- Decapitate mice after perfusion by scissors and peel the skull by forceps.
- Set the cerebrum in the acrylic brain slicer and cut the cerebrum in coronal brain sections (4 mm thick between 1 and 5 mm posterior to bregma) by razor (Figure 2A-C).
- Collect the brain tissue in sampling tubes containing 200 µl of the lysis buffer (Figure 2D).
- Homogenize the brain tissue by Digital Sonifier® (20 kHz, 200 W) (Figure 2E-F).
- Centrifuge the lysates at 15,000 x g for 10 min at 4 °C.
- Collect the supernatant in sampling tubes (Figure 2G).
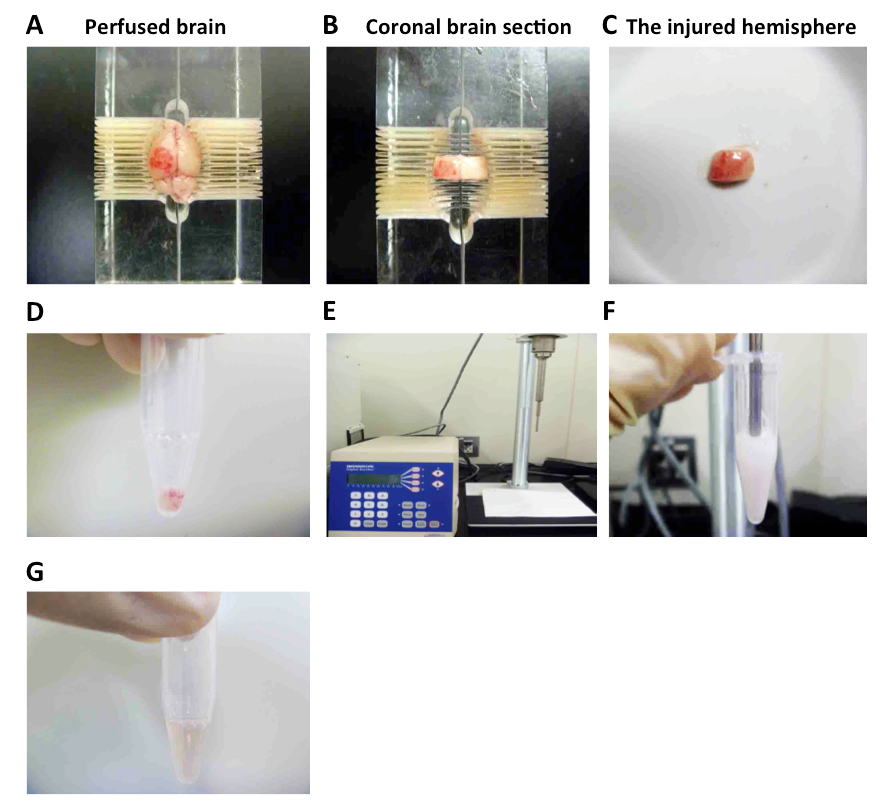
Figure 2. Procedure of mouse perfusion with PBS and collections of perfused cerebrum. A. Perfused mouse cerebrum was isolated and set in the acrylic brain slicer. B. The cerebrum in coronal brain section was cut. C. The injured cortex was isolated from the coronal brain section. D. Collection of the injured mouse cortex. The injured cortex was collected in sampling tubes containing 200 µl of the lysis buffer. E. Digital Sonifier® for homogenization was shown. F. The collected hemispheres were homogenized by Digital Sonifier®. G. After the homogenized samples were centrifuged, the supernatant was collected in sampling tubes.
- Decapitate mice after perfusion by scissors and peel the skull by forceps.
- Measurement of proteins
- Dilute samples 10-fold by lysis buffer.
- Add 20 µl BSA standards (0, 125, 250, 500 and 750 µg/ml) or 20 µl samples to each well in the 96-well plate.
- Mix BCA Protein Assay Reagent A and BCA Protein Assay Reagent B (at rate of 50:1) in a 15 ml conical tube.
- Add 150 µl mixture of BCA Protein Assay Reagent A and BCA Protein Assay Reagent B to each well in the 96-well plate.
- Incubate at room temperature (RT) for 30 min.
- Measure absorbance at OD570 nm by a microplate reader. The protein samples were diluted in 0.2 mg/ml prior to ELISA.
- Dilute samples 10-fold by lysis buffer.
- Preparation of ELISA plate
- Dilute anti-mouse albumin goat antibody in 1/100 with immobilizing solution.
- Add 100 µl of the diluted anti-mouse albumin to each well.
- Seal the plate and incubate it at RT for 4 h.
- Rinse the plate with PBST (200 µl to each well) 3 times.
- Add 200 µl of soaking solution to each well.
- Incubate at RT for 5 min.
- Dry the plate at RT.
- Pack the plate and store at 4 °C at overnight. If you want to finish the ELISA in one day, you can skip this step.
- Dilute anti-mouse albumin goat antibody in 1/100 with immobilizing solution.
- ELISA
- Dilute mouse serum albumin at each concentration (0, 6.25, 12.5, 50 and 100 ng/ml) by lysis buffer. These mouse serum albumin solutions are used as standard solutions.
- Add 50 µl albumin standard solutions or 50 µl samples (10 µg protein) to each well in the prepared ELISA plate.
- Seal the plate and incubate it at RT for 2 h.
- Wash the plate with PBST (200 µl to each well) by using a microplate mixer at medium speed for 5 min 5 times.
- Remove PBST and dry the plate at RT for 15 min.
- Add 100 µl of a HRP-conjugated anti-mouse albumin goat antibody (diluted 1/1,000) with PBS to each well.
- Seal the plate and incubate it at RT for 1 h.
- Wash the plate with PBST (200 µl to each well) by using a microplate mixer at medium speed for 5 min 5 times.
- Add 100 µl of SureBlueTM TMB Microwell Peroxidase Substrate to each well.
- Incubate at RT for 5 min.
- Add 100 µl of 450 nm BioFX® Liquid Nova-Stop Solution for TMB Microwell Substrates to each well.
- Read the plate using a microplate reader at 450 nm.
- Calculate the concentrations of albumin using the standard curve.
- Dilute mouse serum albumin at each concentration (0, 6.25, 12.5, 50 and 100 ng/ml) by lysis buffer. These mouse serum albumin solutions are used as standard solutions.
Representative data
- Albumin contents in the mouse cerebrum after cold injury (with PBS perfusion).
Table 1. Concentrations of albumin standard solutions and their OD450 values. Absorbance of albumin standard solutions was measured at 450 nm. The Albumin standards were duplicated and the averages of duplicated standards were shown.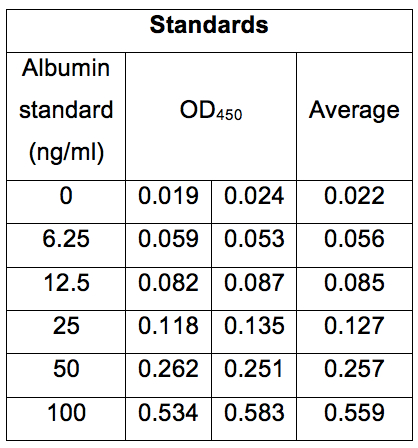
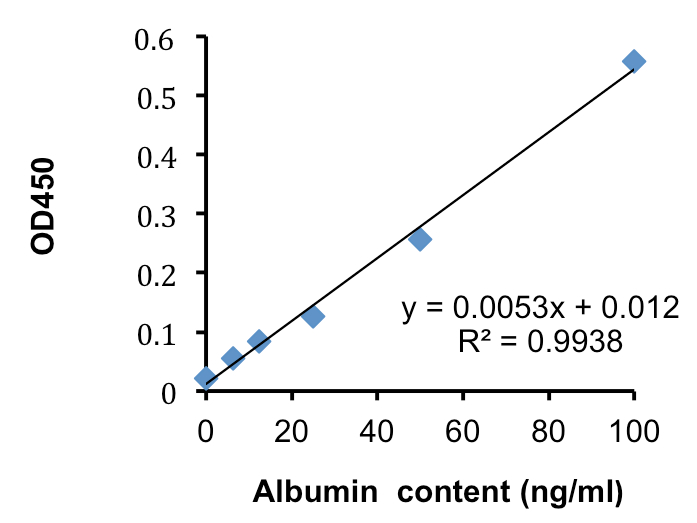
Figure 3. Standard curve of albumin solutions. The standard curve was designed from the OD450 values and the concentrations of albumin standard summarized in Table 1. The vertical axis shows the value of OD450 and the horizontal axis shows albumin standard content.
Table 2. Albumin contents in the perfused mouse cerebrum after cold injury. After cold injury, mice were perfused with PBS. The albumin contents (ng/ml) in mice cerebrum were calculated by equation from the standard curve shown in Figure 3. Brain tissue extracts were diluted at 0.2 mg/ml. Albumin/brain tissue protein (ng/mg) were calculated by dividing albumin contents (ng/ml) by brain tissue protein contents (0.2 mg/ml).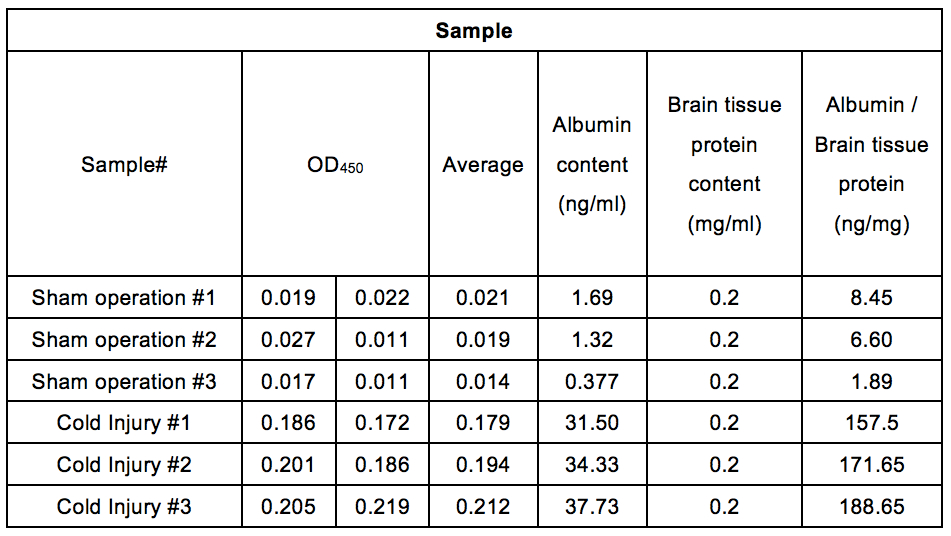
- Albumin contents in the mouse cerebrum after cold injury (without PBS perfusion).
Table 3. Concentrations of albumin standard solutions and their OD450 values. As with Table 1, absorbance of albumin standard solutions was measured at 450 nm.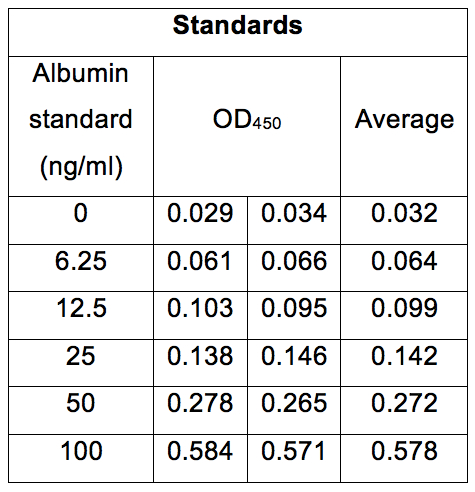
Figure 4. Standard curve of albumin solutions. The standard curve was designed from data in Table 3. The vertical axis shows the OD450 value and the horizontal axis shows albumin standard contents.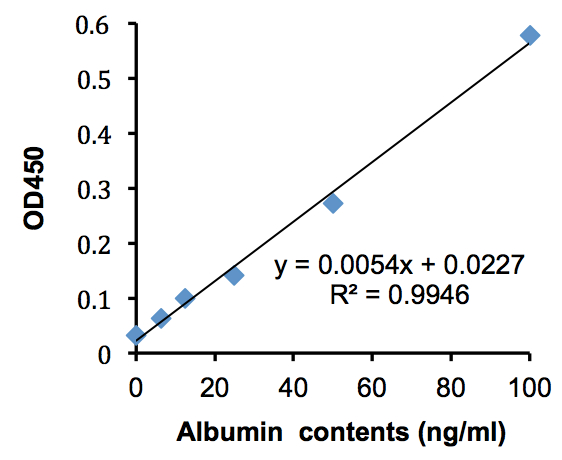
Table 4. Albumin contents in the perfused mouse cerebrum after cold injury (without PBS perfusion). After cold injury, mice cerebrums were collected. The albumin contents (ng/ml) in mice cerebrum were calculated by equation from the standard curve shown in Figure 4. Brain tissue extracts were diluted at 0.2 mg/ml. Albumin/brain tissue protein (ng/mg) were calculated by dividing albumin contents (ng/ml) by brain tissue protein contents (0.2 mg/ml).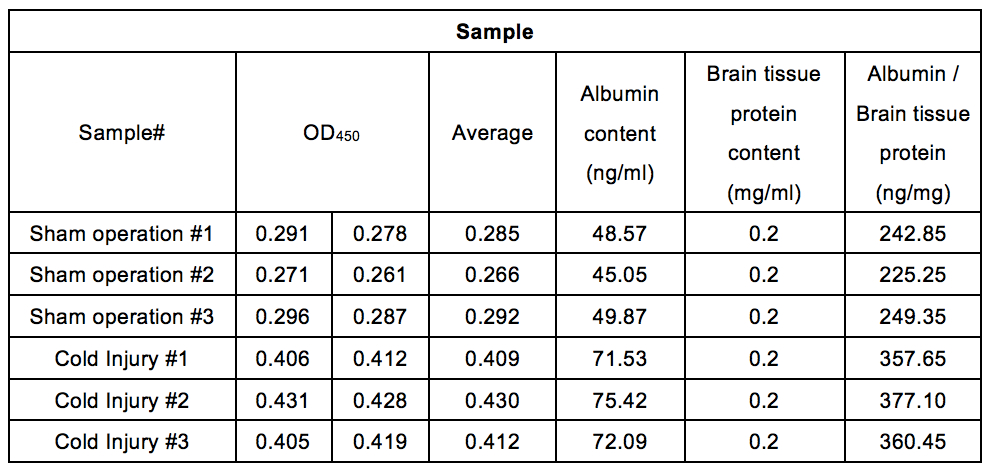
Notes
- In non-injured brain, extravasations of serum proteins are rarely observed. As is indicated above, brain albumin contents of non-injured mouse were reduced to very low levels by perfusion with PBS. This suggests an importance of a successful washing-out of blood components in brain vessels.
Recipes
- PBS
Dilute 10x PBS in distilled water - PBST
PBS
0.05 % Triton X-100 - Lysis buffer
20 mM Tris/HCl (pH 7.4)
Dissolve Tris (hydroxymethyl) aminomethane in distilled water and regulate pH by HCl
150 mM NaCl
1% (v/v) NP-40
0.5% (v/v) deoxycholic acid
0.1% (w/v) SDS
0.5% (w/v) EDTA
2 mM phenylmethylsulfonyl fluoride
10 µg/ml aprotinin
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (C) from the JPSP (21590108).
References
- Michinaga, S., Nagase, M., Matsuyama, E., Yamanaka, D., Seno, N., Fuka, M., Yamamoto, Y. and Koyama, Y. (2014). Amelioration of cold injury-induced cortical brain edema formation by selective endothelin ETB receptor antagonists in mice. PLoS One 9(7): e102009.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Michinaga, S. and Koyama, Y. (2015). ELISA Detection of Endogenous Serum Albumin in the Mouse Brain: A Measure of Extravasation Following Brain Injury. Bio-protocol 5(9): e1469. DOI: 10.21769/BioProtoc.1469.
Category
Neuroscience > Nervous system disorders > Blood brain barrier
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









