- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
FLP/FRT Induction of Mitotic Recombination in Drosophila Germline
Published: Vol 5, Iss 9, May 5, 2015 DOI: 10.21769/BioProtoc.1458 Views: 31302
Reviewed by: Arsalan DaudiTzvetina BrumbarovaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Synthetic Lethality Screens Using RNAi in Combination with CRISPR-based Knockout in Drosophila Cells
Benjamin E. Housden [...] Norbert Perrimon
Feb 5, 2017 11715 Views

Assaying Mechanonociceptive Behavior in Drosophila Larvae
Nina Hoyer [...] Peter Soba
Feb 20, 2018 10090 Views

Assaying Thermo-nociceptive Behavior in Drosophila Larvae
Meike Petersen [...] Peter Soba
Feb 20, 2018 8689 Views
Abstract
The FLP/FRT system is a site-directed recombination technology based on the targeting of a recombination enzyme (flipase - FLP) to specific DNA regions designated as flipase recognition target (FRT) sites. Initially identified in Saccharomyces cerevisiae, the yeast FLP-enzyme and its FRT recombination targets were successfully transferred into each major chromosome arm in Drosophila (Golic and Lindquist, 1989). This offers the ability to mediate mitotic recombination in vivo during development in a controlled manner [revised in Theodosiou and Xu (1998)]. The controlled induction of the mitotic recombination events is usually performed by expressing the FLP under the control of the heat-shock (hs) promoter. This allows the expression of high FLP levels at specific developmental time windows. Strains carrying these genetically marked FLP/FRT chromosomes have greatly enhanced our ability to study gene function in both germline and somatic Drosophila tissues. Here we describe two different protocols: One to induce and identify homozygous mutant clones in ovaries and the other to generate female germline mutants for the analysis of maternal effects on embryogenesis.
Keywords: DrosophilaMaterials and Reagents
- Fly stocks
- w; FRT 42B, fand/CyO (Guilgur et al., 2014)
- y w hs-FLP22/Y; FRT 42B nls-GFP/CyO hs-hid (Guilgur et al., 2014)
- y w hs-FLP22; If/CyO hs hid (Guilgur et al., 2014)
- FRT 42B ovoD/T(1;2)OR64/CyO (Guilgur et al., 2014; Bloomington Drosophila Stock Center, catalog number: 4434)
- w; FRT 42B, fand/CyO (Guilgur et al., 2014)
- Molasses (Sipa Barley Malt48, Provida, catalog number: G109115B )
- Bett syrup (Zuckerrubensirup, catalog number: 01939 )
- Cornmeal (Provida)
- Yeast granulated (LESAFFRE IBÉRICA)
- Soy flour (Salutem, catalog number: 5601557003008 )
- Agar ( Nzytech, catalog number: MB02904 )
- Napagin (Dutscher, catalog number: 789063 )
- Propionic acid (Acros Organics)
- Fly food (see Recipes)
Equipment
- Plastic bottles
- Water bath at 37 °C
- Stereoscope
Procedure
- Generation and labeling of mutant clones using the FLP/FRT system
In this section we describe the generation of homozygous mutant clones for a recessive mutation fandango (fand) which has been characterized in Guilgur et al. (2014). In order to discriminate between fand homozygous mutant and wild-type clones, we combine the FLP/FRT system with a cell-autonomous marker as originally described in Xu and Rubin (1993). We use Green Fluorescent Protein tagged with a nuclear localization signal (nls-GFP) in order to easily identify clones based on the presence (wild type clones-nls-GFP plus) or absence (mutant clones-nls-GFP minus) of a nuclear GFP signal (Figure 1).
Flies were raised using standard techniques at 25 °C unless indicated. All crosses were set up in bottles and flipped to new food vials on a daily basis at least 4 times.- To generate homozygous mosaic clones of fandango allele we crossed virgins carrying the mutant allele (genotype: w; FRT 42B, fand/CyO) with males carrying the following markers: hs-flipase, the FRT 42B, the nls-GFP construct and a balancer CyO bearing the heat-shock-inducible pro-apoptotic transgene head involution defective (hid) (genotype: y w hs-FLP22/Y; FRT 42B nls-GFP/CyO hs-hid). The reverse cross could also be used (see Figure 2).
- By the late second/third larval instar stages the F1 progeny was heat-shocked in a 37 °C water bath for 1 h, in order to induce mitotic recombination and the lethality of organisms carrying the balancer with hid transgene. An additional 1 h heat-shock in the following day could be done to increase the frequency of clones.
Note: The developmental stage selected to induce the mitotic recombination via heat shock can vary depending on the tissue to be analyzed. Doing two consecutive heat shock could increase the rate of recombination as a result of more flipase expression and therefore more clones will be generated. However, it is important be aware about the health of the original stocks used in the cross (sometimes mutant stocks are weak) and more than one heat shock could be harmful for the flies. In our case for the fand allele it was sufficient one heat shock. The number of flies used in the cross referred in section 1 are approximately 15 female virgins with 5 males.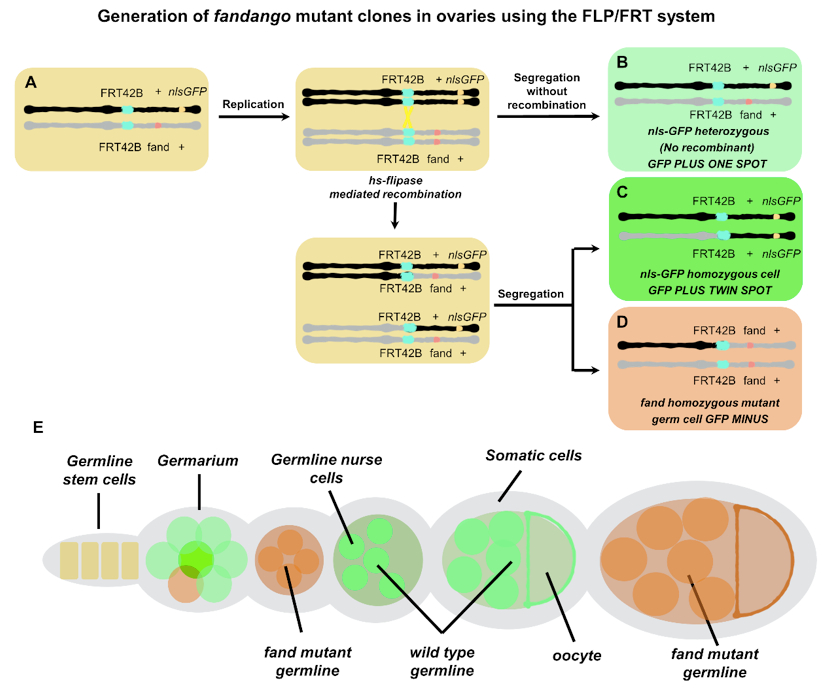
Figure 1. Generating and labeling mutant clones using FLP/FRT system and a cell marker. In a heterozygous parental cell (A), FLP induces mitotic recombination between FRT sites (yellow arrow) on homologous 2R chromosome arms. Segregation of recombinant chromosomes at mitosis produces two daughter cells: A mutant cell bearing two copies of the mutant allele fand (D) and a wild-type cell containing only the wild-type form of the gene (C). The cell marker nls-GFP co-segregates with the wild-type gene (wild type twin-spot clones - nls-GFP plus) and the mutant clone cells are labeling by its absence (fand mutant clones - nls-GFP minus). The non-recombined cells are also identified by the lighter signal from only one copy of nls-GFP (B) (wild type one-spot clones - nls-GFP plus). Schematic representation of clone mosaics in Drosophila ovaries (E). FLP-recombinase target sequences (FRT) are depicted in cyan, fandango mutation in orange and the nls-GFP construct in ochre, heterozygous parental cell in light brown, one-spot cells in light green, twin-spot cells in green, fand mutant clone cells in pink.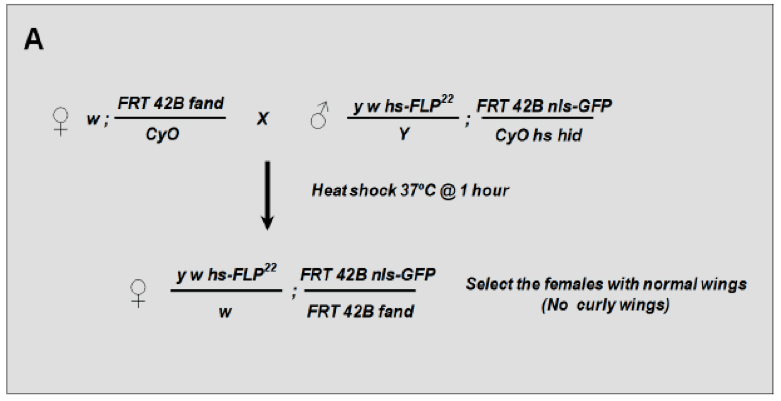
Figure 2. Genetic crosses to recover FRT/FLP induced clones labeled by a cell marker - F1 adult females with normal wings (absence of curly wings indicates lack of the CyO balancer) were transferred to food vials supplemented with fresh baker’s yeast for 3 days prior to ovary dissection and processing (Figure 3A-B).
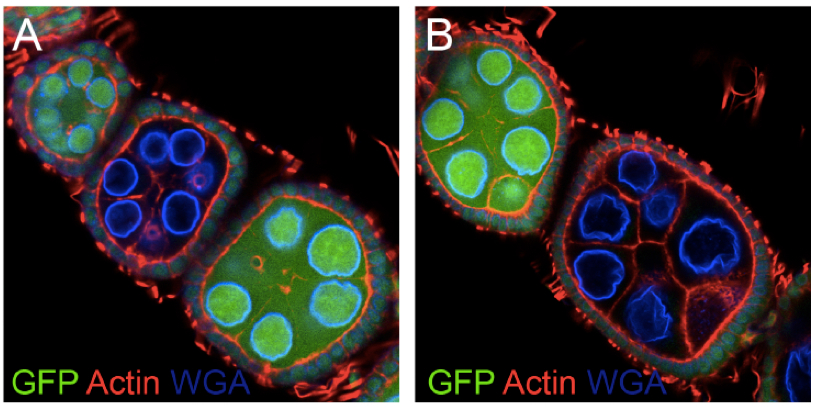
Figure 3. Fandango mutant clones in ovaries. Oogenesis is normal in fandango mutant ovary clones (A, B). Absence of endogenous nls-GFP (nls-GFP minus) indicated that the cells were homozygous for fand mutation. Contrary, presence of endogenous nls-GFP (nls-GFP plus) indicates wild type clones. Ovaries were stained for F-actin (red) and WGA (blue).
- To generate homozygous mosaic clones of fandango allele we crossed virgins carrying the mutant allele (genotype: w; FRT 42B, fand/CyO) with males carrying the following markers: hs-flipase, the FRT 42B, the nls-GFP construct and a balancer CyO bearing the heat-shock-inducible pro-apoptotic transgene head involution defective (hid) (genotype: y w hs-FLP22/Y; FRT 42B nls-GFP/CyO hs-hid). The reverse cross could also be used (see Figure 2).
- Generation of fandango maternal mutant embryos from germline mutant clones using the FLP/FRT OvoD system
In this section we describe the generation of female germline mutant clones to characterize the maternal effect of the recessive zygotic lethal mutation fandango (Guilgur et al., 2014). The genetic technique applied in this assay takes advantage of the properties of the yeast “FLP/FRT” site-specific recombination system in combination with the germline-dependent dominant female sterile OvoD mutation [originally described in Chou and Perrimon (1992)] (Figure 4).
Figure 4. Generating germline mutant clones using FLP/FRT and OvoD system. In a heterozygous parental cell (A), FLP induces mitotic recombination between FRT sites (yellow arrow) on homologous 2R chromosome arms. Segregation of recombinant chromosomes at mitosis produces two daughter cells: a mutant germ cell bearing two copies of the mutant allele fand (D) and a wild-type cell containing only the wild-type form of the gene (C). The presence of germ line-dependent dominant female sterile OvoD mutation blocks oogenesis generating atrophic ovaries (B, C). Therefore, all the developed ovaries are homozygous mutant for fand allele. Schematic representation of the generation of germline clone ovaries during Drosophila oogenesis (E). FLP-recombinase target sequences (FRT) are depicted in cyan, fandango mutation in orange, the OvoD dominant mutation in ochre, heterozygous parental cell in light brown, OvoD mutant cells in light grey, fand mutant clone cells in pink.
Flies were raised using standard techniques at 25 °C unless indicated. All crosses were set up in bottles and flipped to new food vials on a daily basis. The production of germline clones of the fandango allele was based on two crosses:- The objective of the first cross is to generate the so-called “OvoD males” carrying the hs-flipase, the FRT 42B, the dominant female sterile OvoD mutation and a balancer CyO bearing the heat-shock-inducible pro-apoptotic transgene head involution defective (hid) (OvoD males genotype: y w hs-FLP22/Y; FRT 42B OvoD/CyO hs-hid). These males are produced by crossing virgins of the genotype y w hs-FLP22; If/CyO hs hid, with males of the genotype FRT 42B OvoD/T(1;2)OR64/CyO (see Figure 5 cross b1).
Note: These males are easily recognized by the orange color of their eyes and by their curly wings (CyO).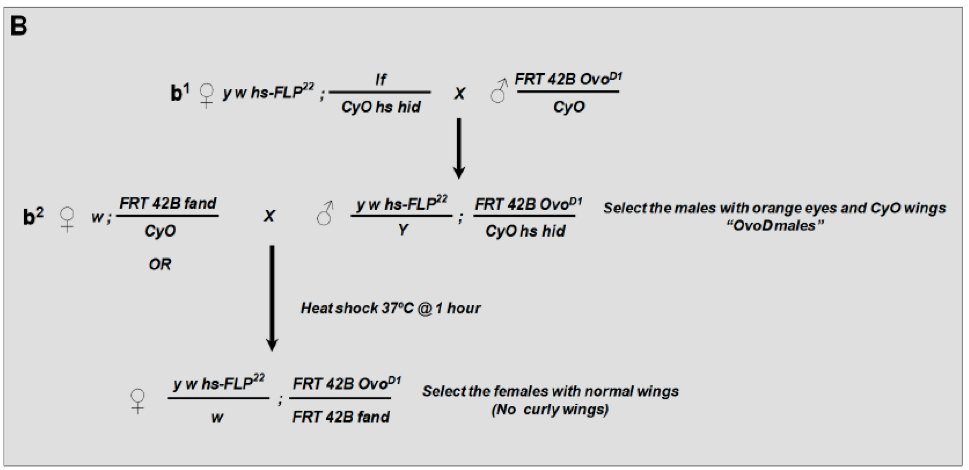
Figure 5. Genetic crosses to recover FRT/FLP induced female flies carrying germline mutant clones - The objective of the second cross is to generate females carrying clonal germline mutants for the fandango allele.
- For this, virgins carrying the fandango mutant allele (genotype: w; FRT 42B, fand/CyO) were crossed with previously produced “OvoD males”.
- By the late second/third larval instar stages, the F1 progeny was heat-shocked in a 37 °C water bath for 1 h, in order to induce mitotic recombination and the lethality of organisms carrying the balancer (see Figure 5 cross b2).
- Finally, F1 adult females with normal wings (lack of the CyO balancer) were transferred to food vials supplemented with fresh baker’s yeast for 3 days prior to egg collection. Since OvoD is a dominant mutation that critically disrupts oogenesis, heterozygous OvoD females develop atrophic ovaries incapable of producing eggs. Accordingly, only the germline stem cells in which FRT-mediated mitotic recombination was induced by the flipase (hs-FLP) will be capable of producing mature eggs. These eggs will necessarily be homozygous for the fandango mutation and can thus be collected, processed and analyzed in order to characterize the embryogenesis phenotypes of the mutation (Figure 6C-D). As controls, germline clones without any associated mutations were generated by crossing virgin flies carrying only the FRT42B recombination site (genotype: w; FRT 42B/CyO) with “OvoD males”, followed by the aforementioned heat shock procedure (Figure 6A-B).
Note: The number of flies used in the cross referred in section 5 are approximately 50 female virgins with 10 males.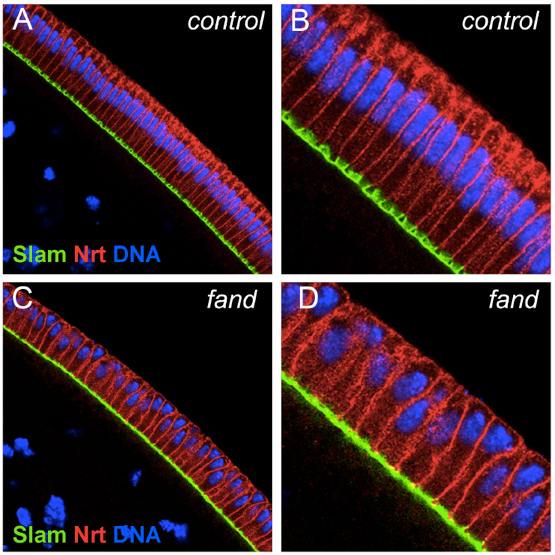
Figure 6. Fandango maternal mutant embryos laid by females carrying germline clones originated by the FRT/FLP OvoD system. Panels show blastoderm cellularized control embryos (hs-FLP; FRT42B), and fand germline clone mutant embryos (hs-FLP; FRT42B fand, maternal mutant) (A-D). Control embryos showed normal epithelial architecture with elongated nuclei and columnar cell shape (A, B). fand germline clone mutant embryos showed abnormal epithelial architecture, the cortical nuclei failed to elongate and became mislocalized (C, D). (B-D) Magnification of C and D respectively. Embryos were stained for Slam (green), Neurotactin (red), and DNA (blue).
- The objective of the first cross is to generate the so-called “OvoD males” carrying the hs-flipase, the FRT 42B, the dominant female sterile OvoD mutation and a balancer CyO bearing the heat-shock-inducible pro-apoptotic transgene head involution defective (hid) (OvoD males genotype: y w hs-FLP22/Y; FRT 42B OvoD/CyO hs-hid). These males are produced by crossing virgins of the genotype y w hs-FLP22; If/CyO hs hid, with males of the genotype FRT 42B OvoD/T(1;2)OR64/CyO (see Figure 5 cross b1).
Recipes
- Fly food (1 L)
Molasses (g): 80
Beet syrup (g): 22
Cornmeal (g): 80
Yeast granulated (g): 18
Soy flour (g): 10
Agar (g): 8
Boiling water (ml): 980
15% Niapagin (ml): 12
Propionic acid (ml): 8
Weigh all ingredients except Niapagin and Proprionic acid in a plastic beaker
Mix all ingredients in a beaker before adding to boiling water
Add boiling water gradually
Transfer the solution to a bottle
Autoclaved at 121 °C, 30 min
When the medium temperature reaches 45 °C-50 °C add Niapagin and Proprionic acid
Acknowledgments
We like to thank Paulo Navarro-Costa for critical reading of manuscript and Rui Martinho for his supervision. Funding: FCT-Fundacao para a Ciencia e Tecnologia (Portugal): Leonardo Gastón Guilgur, SFRH/BPD/47957/2008. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Chou, T. B. and Perrimon, N. (1992). Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131(3): 643-653.
- Golic, K. G. and Lindquist, S. (1989). The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59(3): 499-509.
- Guilgur, L. G., Prudencio, P., Sobral, D., Liszekova, D., Rosa, A. and Martinho, R. G. (2014). Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. Elife 3: e02181.
- Theodosiou, N. A. and Xu, T. (1998). Use of FLP/FRT system to study Drosophila development. Methods 14(4): 355-365.
- Xu, T. and Rubin, G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117(4): 1223-1237.
Article Information
Copyright
Prudêncio and Guilgur. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Prudêncio, P. and Guilgur, L. G. (2015). FLP/FRT Induction of Mitotic Recombination in Drosophila Germline. Bio-protocol 5(9): e1458. DOI: 10.21769/BioProtoc.1458.
- Guilgur, L. G., Prudencio, P., Sobral, D., Liszekova, D., Rosa, A. and Martinho, R. G. (2014). Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. Elife 3: e02181.
Category
Molecular Biology > DNA > DNA recombination
Molecular Biology > DNA > Mutagenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










