- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
DNA Slot Blot Repair Assay
Published: Vol 5, Iss 8, Apr 20, 2015 DOI: 10.21769/BioProtoc.1453 Views: 22070
Reviewed by: HongLok LungPinchas TsukermanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for Quantifying γH2AX Foci in Irradiated Cells Using Immunofluorescence and Fiji Software
Lu Deng [...] Lingying Wu
Aug 20, 2025 2447 Views

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1515 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 223 Views
Abstract
Ultraviolet (UV) irradiation induces helix distorting photolesions such as cyclobutane pyrimidine dimers (CPD) and pyrimidine-pyrimidone (6-4) photoproducts (6-4PP) which threaten genomic integrity if unrepaired. In mammals, nucleotide excision repair (NER) is the only pathway that removes UV-induced DNA damages. Here we describe DNA slot blot repair assay for quantitative detection of NER activity using DNA damage specific antibodies such as anti-CPD and anti-6-4PP. Briefly, genomic DNA irradiated with UV was isolated from cells, and the genomic DNA was vacuum-transferred to a nitrocellulose membrane using a Bio-Dot SF microfiltration apparatus (Bio-Rad). A monoclonal antibody that recognizes CPD or 6-4PP was applied to detect the remaining amount of photolesions in the genomic DNA. For loading control of even loading, DNA onto the membrane can be further analyzed by SYBR-gold staining.
Keywords: DNA repairMaterials and Reagents
- QIAamp® DNA Mini Kit (QIAGEN, catalog number: 51306 )
- Nitrocellulose membrane (GE Healthcare, catalog number: 10600003 )
- Whatman filter paper 3 MM Chr (Whatman, catalog number: 3030-917 )
- Mouse monoclonal antibody to CPD (Kamiya, catalog number: MC-062 )
- Mouse monoclonal antibody to 6-4PPs (COSMO BIO, catalog number: NMDND002 )
- Peroxidase affinipure goat anti-mouse IgG (H+L) (Jackson Laboratories, catalog number: 115-035-003 )
- SYBR® Gold Nucleic Acid Gel Stain (Life Technologies, catalog number: S-11494 )
- RNase A (100 mg/ml) (Bio Basic Canada Inc., catalog number: RB0473 )
- Ethanol (Burdick & Jackson, catalog number: RP090-1 )
- PBS (Bio Basic Canada Inc., catalog number: PD8117 )
- Sodium azide (Junsei Chemical co., catalog number: 26628-22-8 )
- NaOH (Bio Basic Canada Inc., catalog number: SB0617 )
- EDTA (USB Corporation, catalog number: 6381-92-6 )
- Ammonium acetate (NH4Ac) (Katayama Chemical, catalog number: 01-4400 )
- Acetic acid (CH3COOH) (Merck Millipore, catalog number: 64-19-7 )
- NaCl (Bio Basic Canada Inc., catalog number: DB0483 )
- Sodium citrate (Bio Basic Canada Inc., catalog number: CB0035 )
- HCl (Duksan PureChemicals, catalog number: 1129 )
- Tris base (Bio Basic Canada Inc., catalog number: TB0196 )
- Tween-20 (Bio Basic Canada Inc., catalog number: 9005-65-5 )
- Non-fat dry milk (Sigma-Aldrich, catalog number: M7409-1BTL )
- PierceTM ECL Plus Western Blotting Substrate (Life Technologies, catalog number: 32132 )
- 0.4 M NaOH + 10 mM EDTA buffer (see Recipes)
- 2 M ammonium acetate (see Recipes)
- SSC (see Recipes)
- TE buffer (see Recipes)
- PBS-T (see Recipes)
- Blocking buffer (see Recipes)
- Buffer for primary antibody dilution (see Recipes)
- Buffer for secondary antibody (see Recipes)
Equipment
- Ultraviolet irradiator (UVP, catalog number: CL-1000 )
- UV-C sensor (UVP, catalog number: 97-0016-01 )
- Bio-Dot SF microfiltration apparatus (Bio-Rad Laboratories, catalog number: 170-6542 )
- Vacuum pressure pump (Gardner Denver Welch Vacuum Technology Inc., catalog number: 2522C-10 )
- Vacuum drier (JS Research Inc., catalog number: JSVO-30T )
- Orbital shaker (Wisd Laboratory Instruments, catalog number: SHO-1D )
- NanoDrop (Beckman Coulter, catalog number: DU-730 )
- Heat block (Wealtec Corp., catalog number: HB-1 )
- Gel doc (Bio-Rad Laboratories, catalog number: 75S/01085 )
- Centrifuge (Hanil Science Industrial, catalog number: Smart R17 )
- Microcentrifuge tube (SPL, catalog number: 60015 )
- 12-channel multi pipette (HTL LAB Solutions, catalog number: 5127 )
- Paper forcep (USBECK, catalog number: 3110)
- X-ray film 8 x 10 inch (Fujifilm Corporation, catalog number: HR-U30 )
- X-ray Cassette-AFAB TYPE 8 x 10 inch (JPI, catalog number: AFAB8x10 )
Procedure
- Cell culture and UV irradiation
- Cells were allowed to grow confluent in 6-well plates.
- Remove the medium, wash with warming 1x PBS once.
- UV irradiation (10-40 J/m2 of 254 nm UV).
Note: It is necessary to remove the plastic lid before UV irradiation.
- Incubate cells in culture medium for repair for the indicated times before harvest them.
The repair time can be varied depending on the cell’s repair capability. Thus for obtaining the optimal time courses for CPD and 6-4PP repair a pioneer experiment with various time courses should be performed. The following time courses are good to be tested for uncharacterized cells.
Note: General repair time courses for
CPD: No UV, 0, 6, 12, 24, 48 h
6-4PPs: No UV, 0, 1, 2, 4, 8 h.
- Trypsinize and collect each sample in a 1.5 ml microcentrifuge tube by centrifugation at 2,000 x g for 30 sec.
Cell pellets can be stored at deep freezer up to 1 month for later DNA preparation.
- Cells were allowed to grow confluent in 6-well plates.
- DNA preparation (using QIAamp DNA Mini Kit)
- Resuspend cell pellet in 200 μl of 1x PBS (room temperature).
- Add 4 μl of RNase A (100 mg/ml).
- Add 20 μl of QIAGEN proteinase K (or protease).
- Add 200 μl of AL Buffer and Mix by pulse-vortexing for 15 sec.
- Incubate at 56 °C for 10 min.
- Briefly centrifuge to remove drops from the inside of the lid.
- Add 200 μl of EtOH (100%) and Mix again by pulse-vortexing for 15 sec.
- Briefly centrifuge to remove drops from the inside of the lid.
- Apply the mixture to QIAamp Mini spin column (in a 2 ml collection tube) without wetting the rim.
- Close the cap, centrifuge for 1 min at 6,000 x g.
- Place spin column in a clean 2 ml collection tube.
- Add 500 μl of AW1 buffer and centrifuge 6,000 x g, 1 min.
- Place spin column in a clean 2 ml collection tube.
- Add 500 μl of AW2 buffer and centrifuge at full speed (approximately 20,000 x g) for 3 min.
- Place spin column in a clean 1.5 ml E-tube.
- Add 200 μl of AE buffer and Incubate at RT for 5 min and centrifuge 6,000 x g, 1 min.
- Measure DNA concentration using NanoDrop.
- Resuspend cell pellet in 200 μl of 1x PBS (room temperature).
- Slot blotting (using Bio-Dot SF microfiltration apparatus)
- DNA preparation (300 μl total final volume per slot)
- Dilute DNA.
- CPD (50 or 100 ng) → into 150 μl of 0.4 M NaOH + 10 mM EDTA buffer
- 6-4PP (500 ng or 1 μg) →into 150 μl of dH2O (Do not include NaOH buffer)
Note: We used 50 ng or 100 ng of the DNA in 300 μl of total volume to 0.4 M NaOH in 10 mM EDTA for CPD detection. Whereas for 6-4PP detection we used 500 μg or 1 μg of DNA and don't treat the sample with alkali (just use the water) for 6-4PPs since it destroys the 6-4PPs).
- Boil for 10 min at 100 °C to denature the DNA.
- Transfer immediately samples to ice.
- Add 150 μl of cold 2 M ammonium acetate (pH 7.0) into CPD samples to neutralize the DNA (while the DNA is on ice).
- Dilute DNA.
- Slot blotter
- Presoak 3 blotting papers (Whatman filter 3 MM paper) and 1 nitrocellulose membrane in 6x SSC for 10 min.
- Cleaning the blotting Manifold (rinse it in running distilled water).
- Place 3 presoaked blotting papers onto the top of the manifold's middle plate. And then place nitrocellulose membrane on top of the blotting papers.
- Put the slotted portion of the manifold onto top of the membrane.
- Clamp apparatus closed using the two side pieces.

Figure 1. Bio-Dot SF microfiltration apparatus assembly (via Bio-Rad Laboratories)
- Screw the plastic nozzle into the side of the apparatus.
- Connect vacuum line to the nozzle.
- Aspirate excess buffer leaking from presoaked papers and membrane.
- Wash the membrane once with 400 μl of TE into each slot (using multi-channel pipette) and gentle vacuum. We recommend to fill buffer in every well before applying vacuum to deliver steady pressure on each well.
- Carefully load DNA samples into the wells (using 1,000 p pipette) with the vacuum off and the flow valve open.
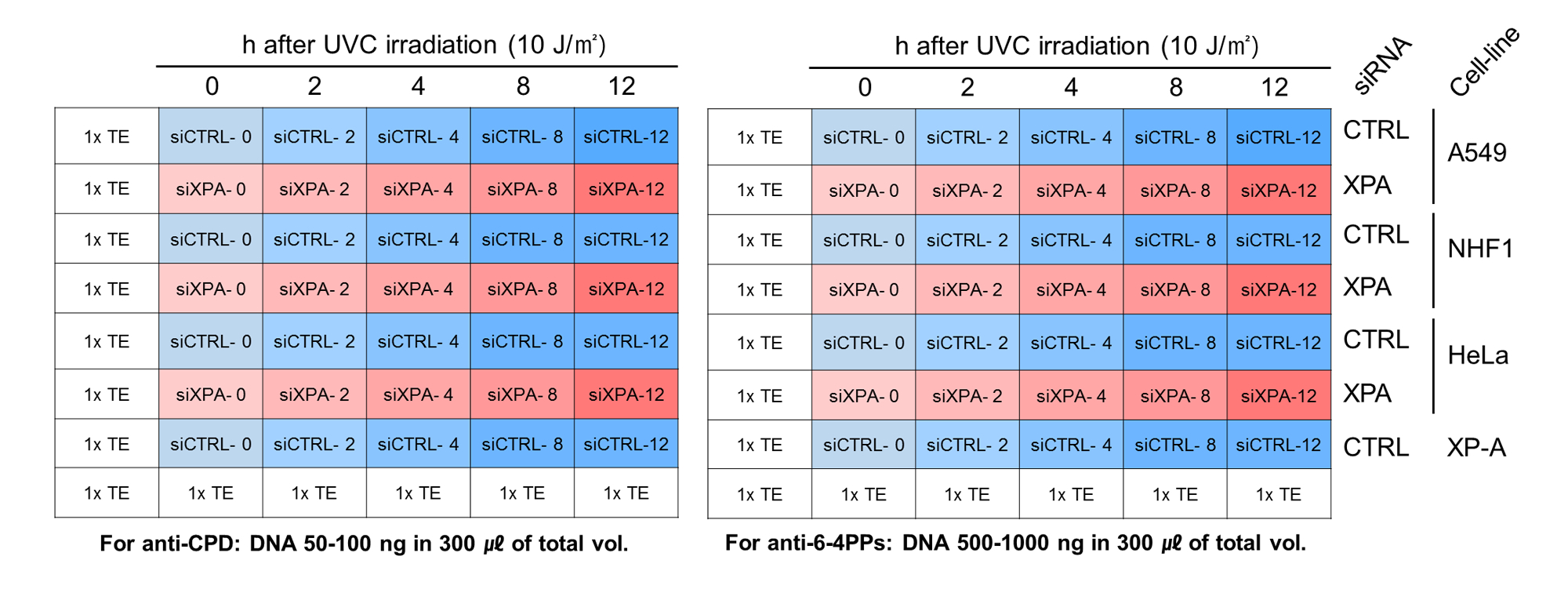
Figure 2. Schematic table of sample loading into the well of blotting Manifold
- Gentle vacuum to attach DNA onto the membrane.
- Add 400 μl of 2x SSC to each slot and gentle vacuum.
- Disassemble the blotting manifold and remove the nitrocellulose membrane and put the membrane between 3 M paper.
- Baking the membrane in the vacuum drier at 80 °C for 2 h.
- Presoak 3 blotting papers (Whatman filter 3 MM paper) and 1 nitrocellulose membrane in 6x SSC for 10 min.
- DNA preparation (300 μl total final volume per slot)
- Immuno-blotting
- Soak the blotting membrane in PBS-T for 10 min with 50 ml PBS-T in the plastic box.
- Transfer the membrane to 30 ml of blocking buffer and shake for 30 min at room temperature.
- Wash the membrane for 3 times (5 min each) with PBS-T.
- Apply the mouse CPD or 6-4PP primary antibody diluted 1:2,000 in PBS-T with 0.02% sodium azide.
- Incubate the blot in a cold room with shaking for overnight (12-14 h).
- Wash the membrane with PBS-T 4 times for 15 min each.
- Incubate with secondary antibody (anti-mouse IgG HRP-conjugated) diluted 1:2,500 in blocking buffer for 1 h at room temperature on an orbital shaker.
- Wash the membrane with PBS-T 4 times for 15 min each.
- Apply ECL and expose to film.
- Soak the blotting membrane in PBS-T for 10 min with 50 ml PBS-T in the plastic box.
- DNA staining
- Rinse the membrane with PBS-T.
- Incubate the membrane with 10 ml of PBS-T containing SYBR-gold dye diluted 1:10,000 in PBS-T for 20 min at room temperatures on an orbital shaker.
- Wash the membrane with PBS-T 2 times for 5 min each.
- Take a photo of SYBR-gold stained blot under UV transilluminator.
Note: The DNA bound to the membrane was detected with SYBR-Gold dye and used as loading control.
- Rinse the membrane with PBS-T.
Representative data

Figure 3. Representative data for slot blot repair assay. XPA is a key factor of nucleotide exision repair for UV-induced DNA damage. Knock down of XPA using siRNA affected the repair of both CPD and the (6-4) PP. Control siRNA target cyclophilin B housekeeping gene. (A and B) Reduction in XPA level proportionally affected the repair of both CPD and the (6-4) PP as measured by slot–blot assay. Genomic DNA from cells irradiated with 10 J/m2 of UV was analyzed to detect residual CPD (A) or (6-4) PP (B) during the recovery times as indicated. C. Protein levels in human cells were analyzed by immunoblotting with the indicated antibodies from cells transfected with 100 nM siRNA targeting XPA orcyclophilin B (Control, CTRL). This figure was adapted from Kang et al. (2011).
Recipes
- 0.4 M NaOH + 10 mM EDTA buffer (500 ml)
NaOH 8 g
0.5 M EDTA 10 ml
Adjust volume to correct volume with dH2O
- 2 M ammonium acetate (pH 7) (500 ml)
Ammonium acetate (NH4Ac) 77.08 g
Mix in ~400 ml dH2O, adjust pH to 7 with acetic acid (CH3COOH), then adjust volume to 500 ml
Stored at 4 °C
- SSC (pH 7)
- 20x SSC (pH 7) (1 L)
NaCl 175.3 g
Sodium citrate 88.2 g
Mix in ~ 700 ml dH2O, adjust pH to 7 with HCl, then adjust volume to 1 L
- 2x SSC (1 L)
20x SSC 100 ml
dH2O 900 ml
- 6x SSC (1 L)
20x SSC 300 ml
dH2O 700 ml
- 20x SSC (pH 7) (1 L)
- TE buffer
- 1 M Tris-HCL buffer (pH 7.5) 1 L
Mix 121.1 g Tris with 800 ml dH2O
pH to 7.5 with HCl and add dH2O to 1 L
- 10x TE
1 M Tris-HCl (pH 7.5) 100 ml
0.5 M EDTA 20 ml
dH2O 880 ml
- 1x TE
10x TE 100 ml
dH2O 900 ml
- 1 M Tris-HCL buffer (pH 7.5) 1 L
- PBS-T
PBS with 0.1% Tween-20
- Blocking buffer (50 ml)
Mix 2.5 g non-fat dry milk with 50 ml 1x PBS-T
- Buffer for primary antibody dilution
Primary antibody (mouse monoclonal antibody to CPD and 6-4PP) diluted 1:2,000 in PBS-T with 0.02 % sodium azide
- Buffer for secondary antibody
Secondary antibody (anti-mouse IgG HRP-conjugate) diluted 1:2,500 in blocking buffer
Acknowledgments
This work was supported by a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1420070) and National Research Foundation (NRF) of Republic of Korea (NRF-2011-0013804 and NRF-2013-R1A2A2A04008115), and Brain Busan 21 Project.
References
- Kang, T. H., Reardon, J. T. and Sancar, A. (2011). Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res 39(8): 3176-3187.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Park, J. and Kang, T. (2015). DNA Slot Blot Repair Assay. Bio-protocol 5(8): e1453. DOI: 10.21769/BioProtoc.1453.
Category
Molecular Biology > DNA > DNA damage and repair
Molecular Biology > DNA > DNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









