- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Nucleotide Triphosphate Sugar Transferase Activity via Generation of Pyrophosphate
Published: Vol 5, Iss 8, Apr 20, 2015 DOI: 10.21769/BioProtoc.1450 Views: 10412
Reviewed by: Claudia CatalanottiKanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analytical Gel Filtration for Probing Heavy Metal Transfer between Proteins
Steffen Lorenz Drees and Mathias Lübben
Aug 5, 2016 11109 Views

Hydrogen Deuterium Exchange Mass Spectrometry of Oxygen Sensitive Proteins
Luke Berry [...] Brian Bothner
Mar 20, 2018 10258 Views

Binding Affinity Quantifications of the Bacteriophage Mu DNA Modification Protein Mom Using Microscale Thermophoresis (MST)
Shubha Udupa [...] Shweta Karambelkar
Jul 20, 2022 3707 Views
Abstract
Nucleotide triphosphate (NTP) transferases (EC. 2. 7. 7. X) transfer a nucleoside monophosphate moiety from NTP to another substrate. NTP sugar transferases form a large member of the NTP transferase. There are many variations for the substrate combination of the NTP sugar transferases. It is important to measure the precise enzymatic activity of such NTP sugar transferases by a simple and efficient method. In our method, we measure pyrophosphate as a byproduct of nucleotide diphosphate (NDP)-sugar generation using the pyrophosphate assay kit. The kit reagents include two enzymes that convert pyrophosphate to phosphate, and then phosphorolyze chromogenic substrate to allow color development at 360 nm (see details below). Thus, the NDP-sugar formation can be simply traced as production of pyrophosphate, which is monitored by absorbance at 360 nm. This method is reliable and versatile for measurements of various pyrophosphate-producing enzymes that include NTP sugar transferases.
[Principle and overview] NTP transferases catalyze the reversible reaction as follows: NTP + sugar-1P <-> NDP-sugar + PPi
The enzyme reaction can be monitored as generation of inorganic pyrophosphate (PPi). The EnzChek Pyrophosphate Assay kit (Molecular Probes, Life Technologies, Carlsbad, CA) includes two enzymes and sufficient materials for color development to quantitate pyrophosphate. The inorganic pyrophosphatase (component E in the kit) degrades pyrophosphate into phosphate. Purine nucleoside phosphorylase (PNP, component B) utilizes phosphate to cleave the colorgenic substrate 2-amino-6-mercapto-7-methylpurine ribonucleoside (MESG, component A) into ribose-1-phosphate and 2-amino-6-mercapto-7-methylpurine. The product 2-amino-6-mercapto-7-methylpurine has the absorption maximum at 360 nm. The component C is the dilution solution that includes minimal MgCl2 sufficient for the inorganic pyrophosphatase. Thus, NTP transferase activity can be monitored at 360 nm as generation of a byproduct pyrophosphate.
Typically, the nucleotidyl sugar transferase reactions have been measured by High Performance Liquid Chromatography (HPLC) (Kawano et al., 2014). There are several advantages and disadvantages in HPLC method and our enzymatic method (O: advantage, X: disadvantage).
{HPLC method}
O) Can measure the reaction in both directions (NDP-sugar formation and degradation)
O) Can measure with a small amount of the sample protein
X) Can not follow the real-time reaction
X) The peaks of substrates and products must be separated in the chromatogram
{Pyrophosphate assay method}
O) Can observe the reaction in real-time
O) Easy to use many kinds of substrates because of the simple detection at 360 nm
O) Can be used in other pyrophosphate or phosphate generating reactions such as adenylate cyclase, diguanylate cyclase ( Enomoto et al., 2014), and DNA polymerase
X) Can not measure the pyrophosphate-consuming direction of the reversible reaction of NDP-sugar pyrophosphorylase. Km and kcat for NDP-sugar and pyrophosphate are not obtained by the pyrophosphate assay method but by HPLC method.
X) Need certain amount of the experimental protein. The maximum activity that the kit reagents allow corresponds theoretically to the rate of color development for the positive control is 2 x 10-2 U in our case. The instruction states the minimum detection of 5 x 10-5 U. Of course, the minimum activity we can measure may depend on the background activity due to contaminants in the substrates or in the sample preparation.
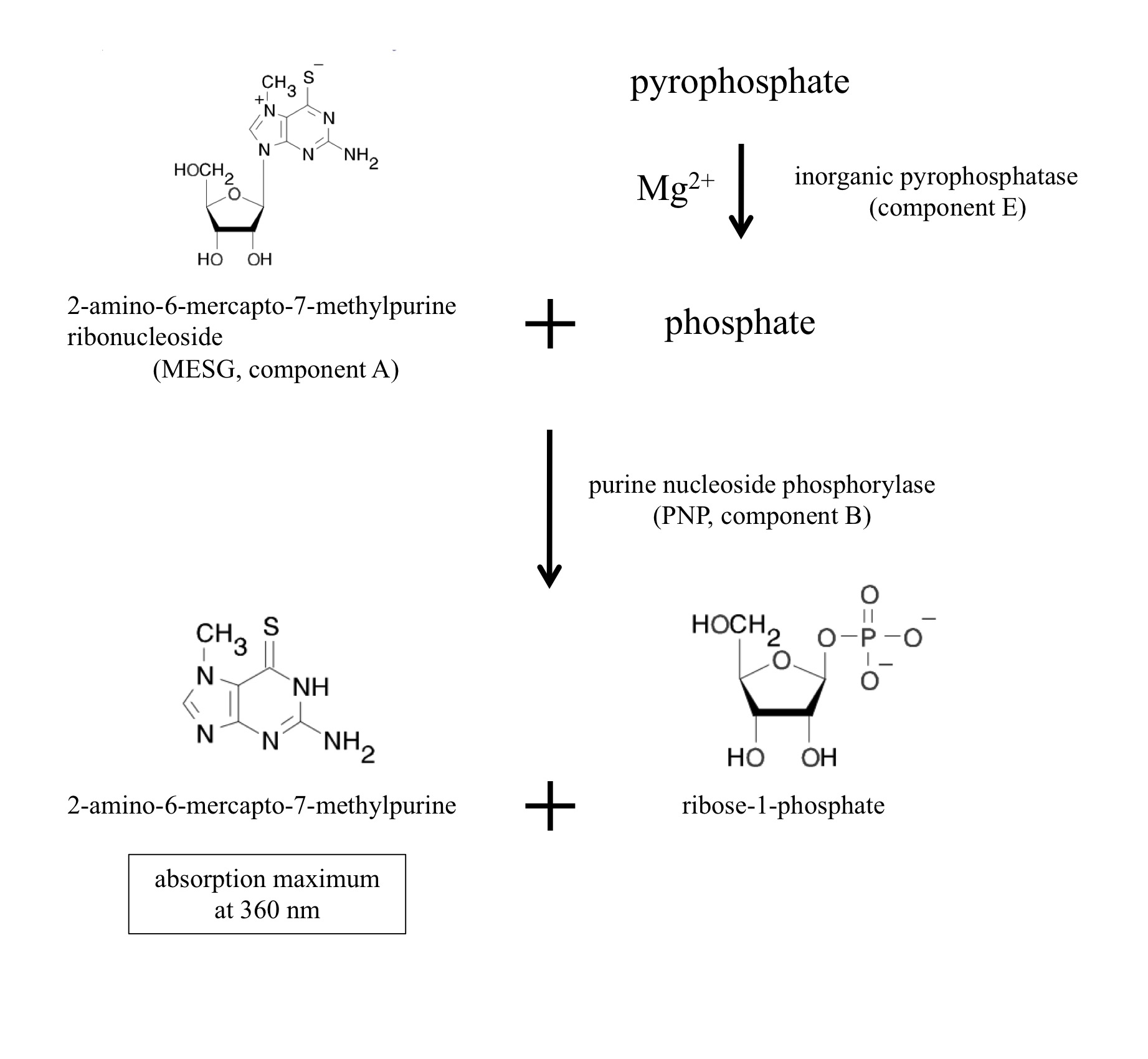
Figure 1. Chromogenic reactions in the assay kit
Materials and Reagents
- EnzChek Pyrophosphate Assay kit (Molecular Probes, catalog number: E-6645 )
- The contents of the components A~E are shown below.
- 2-amino-6-mercapto-7-methylpurine ribonucleoside (MESG)
- Purine nucleoside phosphorylase (PNP)
- 20x reaction buffer: 1.0 mM Tris-HCl, 20 mM MgCl2 (pH 7.5)
- Pyrophosphate standard solution: 50 mM Na4P2O7
- Inorganic pyrophosphatase
- The contents of the components A~E are shown below.
- 1 M MgCl2
- 100 mM substrates (our examples in Table 1)
- Enzyme preparation (with or without imidazole, see note 5 below)
Equipment
- Block incubator (ASTEC, model: BI-516S )
- Spectrophotometer (Shimadzu, model: UV-2600 )
- Electronic cooling and heating cuvette holder (Shimadzu, model: S-1700 )
- Quarts cuvette with a lid
Table 1. An example of substrate reagentsSubstrate Company Catalog number Adenosine triphosphate (ATP) Sigma-Aldrich A-2383 Guanosine triphosphate (GTP) Sigma-Aldrich G-8877 Cytidine triphosphate (CTP) Sigma-Aldrich C-1506 Uridine triphosphate (UTP) Sigma-Aldrich U-6625 Deoxythymidine triphosphate (dTTP) Sigma-Aldrich T-0251 Glucose-1-phosphate (Glc-1P) Sigma-Aldrich G-9380 N-acetylglucosamine-1-phosphate (GlcNAc-1P) Sigma-Aldrich A-2142 Galactose-1-phosphate (Gal-1P) Sigma-Aldrich G-0380 Mannose-1-phosphate (Man-1P) Santa Cruz
BiotechnologySC-284868
Procedure
- Measurement of enzyme activity
- Set up the spectrophotometer and electronic cooling and heating cuvette holder. Hold a cuvette in the holder to keep the temperature (usually 37 °C).
- Prepare the assay kit solutions as written in the product manual and keep them on ice. The instruction states component A should be used within 4 h when melted.
- Prepare reaction mixtures according to Table 2.
- Preincubate the mixture at 22 °C for 20 min to react the contaminated pyrophosphate and phosphate in the block incubator. We usually incubate the mixture for another 10 min at 37 °C (the measuring temperature) to warm up the mixture. The original instruction recommends the preincubation for 10 min at 22 °C.
- Put the reaction mixture into the cuvette in the spectrophotometer and record the absorbance at 360 nm in a time-scan mode for 5 ~ 10 min. Usually, the linear time course of the color development was obtained often initial lag time of 10 ~ 20 sec. Therefore, it may be sufficient to record time points every 30 ~ 60 sec.
- Set up the spectrophotometer and electronic cooling and heating cuvette holder. Hold a cuvette in the holder to keep the temperature (usually 37 °C).
- Data analysis
- Measure the rate of absorbance change (∆A360/sec).
- Convert the reaction rate (∆A360/sec) to the velocity (molar concentration/sec) by the difference of final A360 between the positive control and the negative control 1 (see the formula below).
V (M / sec) = ∆A360 (cm-1) / sec - ∆A360 (cm-1) / sec (negative control) ε (M-1 cm-1)
We experimentally determine the molar extinction coefficient of ~ 11,000 (M-1 cm-1) at 360 nm every time based on the final absorption of the positive control as follows.ε (M-1 cm-1) = final A360 (cm-1, positive control) - final A360 (cm-1, negative control 1) pyrophosphate concentration (M, positive control)
The kit provides standard solution of 50 mM pyrophosphate (component D). We obtained fairy reproducible value for ε in our hands although the instruction does not mention about the known ε value, the original literature (Upson et al., 1996) states ε (M-1 cm-1) = 11,000 at pH 7.6.
Table 2. Preparation of reaction mixtures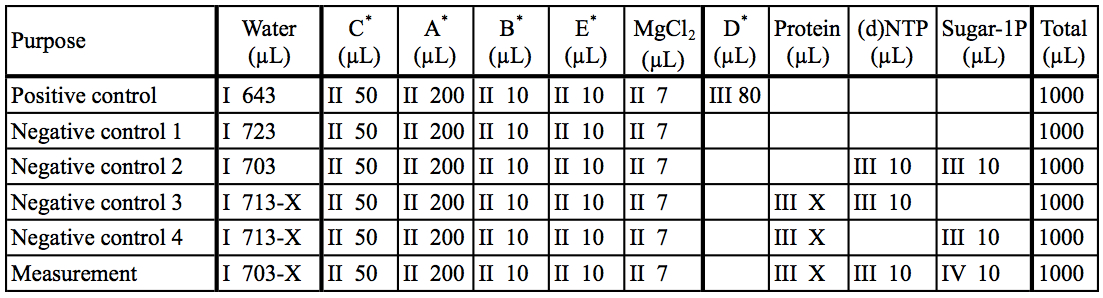
The asterisk represent the kit components A~E. I~IV: The order of addition for preparation of reaction mixtures.
- Measure the rate of absorbance change (∆A360/sec).
Representative data
Here are representative data of cyanobacterial UDP-glucose pyrophosphorylase expressed and purified from E. coli cells (Maeda et al., 2014). The initial slope of the graph was used for calculation. We usually use enzymes at 50 ~ 200 nM, and substrates [(d)NTP and sugar-1P] at 50 µM ~ 2 mM.
Table 3. Preparation of reaction mixtures in our example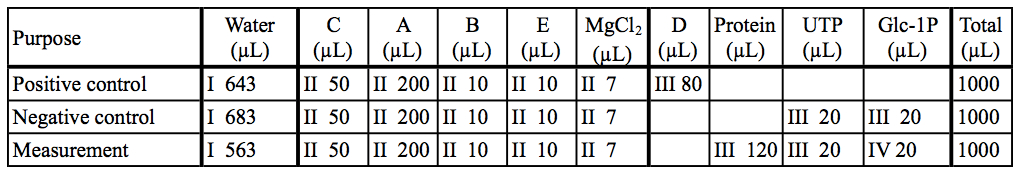
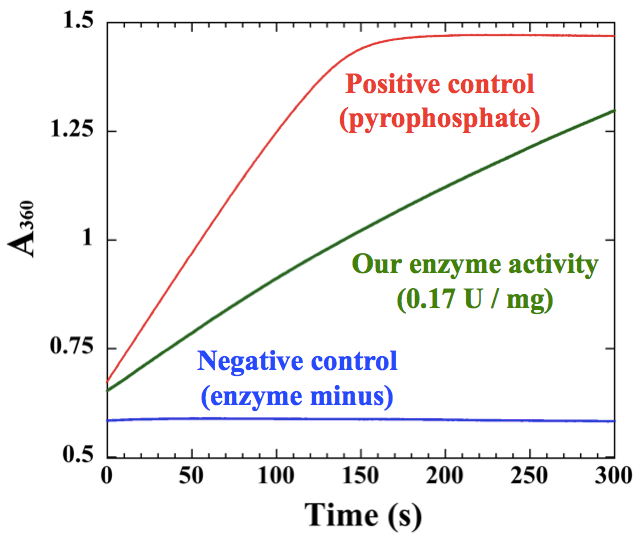
Figure 2. An example of measurements for polyhistidine-tagged cyanobacterial UDP-glucose pyrophosphorylase, which was purified by Ni-affinity chromatography
Notes
- Before measurement, it is essential to check the reaction capacity of kit. Add 50~75 µM sodium pyrophosphate to the kit to confirm the maximum activity of pyrophosphorylase and purine nucleoside phosphorylase in the kit. We cannot measure NTP transferase activities higher than the maximum activity of the kit (Figure 2).
- To check the quality of the assay kit, we must measure the positive control first.
- Negative control 2 gives the level of contamination of phosphate and pyrophosphate. Negative control 3 and 4 give background enzyme activities to degrade (d)NTP and sugar-1P, respectively. Phosphate and pyrophosphate in the sample solutions or substrate reagents increase the background absorbance at 360 nm. We experienced some reagents such as mannose-1-phosphate contained too much phosphate. Such impurities may depend on the lot of the reagent or product manufacturer.
- A simple representative example for an enzyme sample is shown in Figure 2. The slope of the sample should be between the pyrophosphate positive control and the negative control. The initial slope of the graph was used for calculation according to the Data analysis.
- The imidazole (up to final 100 mM in the assay mixture) has little effect on the assay kit activity. This means that the kit accepts usual preparations of His-tagged enzymes, which are (partially) purified by Ni-affinity chromatography.
- Manganese ion (Mn2+) inhibits the assay kit activity at final 5 mM or even lower concentration.
- Reproducibility of the measurements is pretty high, though the maximum activity of the kit depends on the freshness of the kit.
- According to the kit instructions, the reagents of the kit are stable for six months to one year at < -20 °C. Reconstituted MESG (component A) may be stored at < -20 °C for at least one month. Reconstituted purine nucleoside phosphorylase (component B) may be stored at 4 °C for at least one month. Diluted inorganic pyrophosphatase (component E) may be stored at 4 °C for at least one week.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from MEXT and PRESTO from JST (to R. N.) and by Grants-in-Aid for Scientific Research and the GCOE program “From the Earth to Earths” from MEXT, and CREST from JST (to M. I.).
References
- Enomoto, G., Nomura, R., Shimada, T., Ni Ni, W., Narikawa, R., and Ikeuchi, M. (2014). Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. J Biol Chem 289(36): 24801-9.
- Kawano, Y., Sekine, M. and Ihara, M. (2014). Identification and characterization of UDP-glucose pyrophosphorylase in cyanobacteria Anabaena sp. PCC 7120. J Biosci Bioeng 117(5): 531-538.
- Maeda, K., Narikawa, R., and Ikeuchi, M. (2014). CugP is a novel ubiquitous non-GalU-type bacterial UDP-glucose pyrophosphorylase found in cyanobacteria. J Bacteriol 196 (13): 2348-54.
- Upson, R. H., Haugland, R. P., Malekzadeh, M. N. and Haugland, R. P. (1996). A spectrophotometric method to measure enzymatic activity in reactions that generate inorganic pyrophosphate. Anal Biochem 243(1): 41-45.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Maeda, K., Narikawa, R. and Ikeuchi, M. (2015). Measurement of Nucleotide Triphosphate Sugar Transferase Activity via Generation of Pyrophosphate. Bio-protocol 5(8): e1450. DOI: 10.21769/BioProtoc.1450.
Category
Biochemistry > Protein > Interaction > Protein-ligand interaction
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










