- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Introduction and Sequencing of Patient-isolated HBV RT Sequences into the HBV 1.2-mer Replicon
Published: Vol 5, Iss 8, Apr 20, 2015 DOI: 10.21769/BioProtoc.1449 Views: 9697
Reviewed by: Yannick DebingAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro DNA Polymerization Activity Assay Using Cell-free Extracts
Anurag K. Sinha and Malay K. Ray
Aug 20, 2014 9250 Views

Estimation of the Minimum Number of Replication Origins Per Chromosome in any Organism
Marcelo S. da Silva
Oct 20, 2020 4215 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3038 Views
Abstract
Antiviral agents for the suppression of hepatitis B virus (HBV) have been used for treating chronic hepatitis B. However, the emergence of drug-resistant HBV is still a major problem for antiviral treatment. To identify and characterize the drug-resistant HBV, the construction of HBV replicon and in vitro drug susceptibility assay are essential. Here we describe the experimental methods to study drug-resistant HBV.
Keywords: Hepatitis B virusMaterials and Reagents
- Sera of patients with chronic hepatitis B (100~200 μl)
- QIAamp MinElute virus spin kit (QIAGEN, catalog number: 57704 )
- Ex taq polymerase (Takara Bio Company, catalog number: RR001A )
- Forward primer (XhoI site is underlined): 5’-AAT CTT CTC GAG GAC TGG GGA CCC TGC ACC-3’
- Reverse primer (NcoI site is underlined): 5’-GAG CAG CCA TGG GAA GGA GGT GTA TTT CCG-3’
- Gel/PCR DNA Extraction kit (Real Biotech Corporation, catalog number: YDF100 )
- pGEM-T Easy vector (Promega Corporation, catalog number: A1360 )
- T4 ligase (Takara Bio Company, catalog number: 2011A )
- LB broth high salt (Duchefa Biochemie, catalog number: L1704 )
- MacConkey agar (BD, catalog number: 212123 )
- LaboPass Plasmid Mini Purification Kit (COSMO Genetech, catalog number: CMP0111 )
- HBV WT complete sequence (genotype C, GQ872210)
- Xho I (New England BioLabs, catalog number: R0146S )
- Nco I (New England BioLabs, catalog number: R0193S )
- CutSmart™ buffer (New England BioLabs, catalog number: B7204S )
- HBV 1.2 replicon (Figure 1)
- TE buffer (see Recipes)
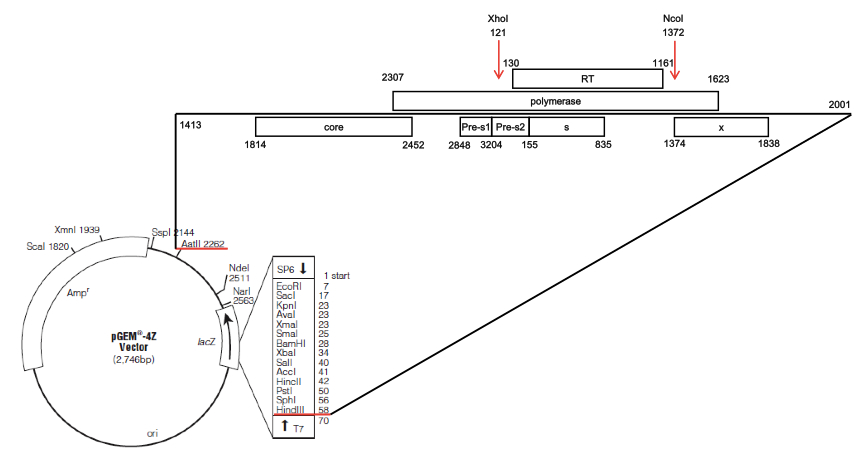
Figure 1. A plasmid map of the HBV 1.2-mer replicon with XhoI and NcoI sites highlighted
Equipment
- Centrifuge (Eppendorf, catalog number: 5415R )
- Pipet Aid XP (Drummond Scientific Company, catalog number: 4-000-202-E )
- MJ Mini 48-Well Personal Thermal Cycler (Bio-Rad Laboratories, catalog number: PTC-1148 )
- Nanodrop Spectrophotometer (Nanodrop, catalog number: ND-1000 )
- Heat block
Procedure
- Amplification of HBV RT domain from patients’ sera
- Prepare 100~200 μl of patient’ serum.
- Extract the HBV DNA using QIAamp MinElute virus spin kit according to the manufacturer’s protocol and elute viral DNA in 50 μl of elution buffer (provided in the kit) or distilled water.
- Amplify HBV RT domain with primers following composition.
HBV DNA (50 ng/μl) 1 μl Forward primer (10 pM) 1 μl Reverse primer (10 pM) 1 μl 10x Ex Taq polymerase buffer 2 μl dNTP mixture (2.5 mM) 2 μl Ex Taq polymerase (5 U/μl ) 0.2μl Distilled water 12.8 μl Total 20 μl - Run PCR as follows: 95 °C for 5 min, followed by 30 cycles of 95 °C for 50 sec, 62 °C for 50 sec, 72 °C 1 min 20 sec, and final extensions at 72 °C for 10 min.
- Identify the PCR products (HBV RT domain) on 1% agarose gel by electrophoresis (Figure 2) and purify with Gel/PCR DNA Extraction kit according to the manufacturer’s protocol (final elution volume is 30 μl of TE buffer).
- Prepare 100~200 μl of patient’ serum.
- Sequencing of HBV RT domain
- Mix the ligation reactions in 1.5 ml tube as described below and incubate overnight at 4 °C.
Purified PCR products (60 ng/μl ) 1 μl pGEM-T Easy vector (50 ng/μl ) 5 μl 2x Rapid Ligation buffer 0.3 μl T4 DNA ligase (3 Weiss units/μl ) 2.7 μl Distilled water 1 μl Total 10 μl - To inactivate the T4 ligase, incubate the ligation reactions at 65 °C for 15 min and place in ice.
- During step B2, prepare the E.Coli (DH5α) competent cells from -80 °C storage and place in ice until thawed.
- Transfer 50 μl of competent cells into ligation reactions and incubate in ice for 15 min.
- Heat-shock the competent cells for 90 sec in heat block at 42 °C, then immediately incubate in ice for 15 min.
- Add 500 μl LB medium to tube from step B5 and incubate for 2 h at 37 °C with shaking.
- Centrifuge the tube at 3,000 rpm for 5 min at room temperate, and then remove the 350 μl of supernatant.
- Gently mix and plate the residue supernatant (approximate 150 μl) onto Ampr MacConkey agar plates using spreader.
- Incubate the plates overnight at 37 °C and check the number of white colonies (approximately 100 colonies should be observed).
- Pick the white colonies, then transfer into 2 ml of LB medium with ampicillin in 15 ml tube and incubate overnight at 37 °C with shaking.
- Extract the DNA plasmids contained the HBV RT domain from bacterial culture (step B10) using the LaboPass Plasmid Mini Purification Kit following the manufacturer’s protocol and elutes plasmids in 50 μl of EB buffer (provided in Kit) or TE buffer.
- Confirm the correct insert DNA by simultaneous digestion reactions for 2 h at 37 °C as described below.
Plasmid DNA (200 ng) - XhoI (20 U/μl ) 0.25 μl NcoI (10 U/μl ) 0.25 μl 10x CutSmartTM Buffer 1 μl Distilled water up to 10 μl Total 10 μl - Sequencing the HBV RT domain by T7 (5’-TAA TAC GAC TCA CTA TAG GG-3’) and SP6 (5’- ATT TAG GTG ACA CTA TAG-3’) promoter primers (located in pGEM-T Easy vector) and analyze the mutations compared with WT HBV (genotype C, GQ872210).
- Mix the ligation reactions in 1.5 ml tube as described below and incubate overnight at 4 °C.
- Cloning of HBV RT domain into HBV 1.2 replicon
- Simultaneously digest 200 ng of plasmids [HBV RT plasmid (step B12) for insert DNA and HBV 1.2mer replicon for vector DNA] with digestion reactions (see the procedure C step 12).
- Identify 1.2 kb (insert DNA, step B12) and 4.8 kb (vector DNA) fragment on 0.8% agarose gel by electrophoresis (Figure 3) and purify the digested DNAs with Gel/PCR DNA Extraction kit according to the manufacturer’s protocol (final elution volume is 30 μl of TE buffer).
- Mix the ligation reactions in 1.5 ml tube as described below and incubate overnight at 4 °C.
Insert DNA (40 ng/μl) 1 μl Vector DNA (50 ng/μl) 1 μl 10x T4 ligase buffer 1 μl T4 DNA ligase (3 Weiss units/μl) 0.3 μl Distilled water 6.7 μl Total 10 μl - Transform (see steps B2~7) and plate onto Ampr LB agar plates.
- Isolate and confirm the plasmid DNA (see steps B9~13).
- Simultaneously digest 200 ng of plasmids [HBV RT plasmid (step B12) for insert DNA and HBV 1.2mer replicon for vector DNA] with digestion reactions (see the procedure C step 12).
Representative data
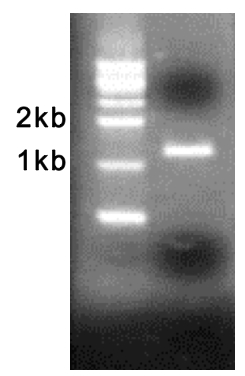
Figure 2. Representative data of amplified HBV RT gene. Prepared HBV DNA from patients sera were used as template. The product size is approximately 1.2 kb.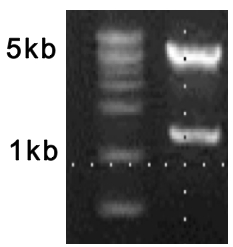
Figure 3. Confirmation of HBV 1.2-mer replicon. Completed HBV 1.2mer replicon was digested with XhoI, NcoI restriction enzyme and was separated on a 1% agarose gel.
Recipes
- TE buffer
10 mM Tris (pH 8.0)
1 mM EDTA
Acknowledgments
This study was supported by Konkuk University.
References
- Ahn, S. H., Park, Y. K., Park, E. S., Kim, J. H., Kim, D. H., Lim, K. H., Jang, M. S., Choe, W. H., Ko, S. Y., Sung, I. K., Kwon, S. Y. and Kim, K. H. (2014). The impact of the hepatitis B virus polymerase rtA181T mutation on replication and drug resistance is potentially affected by overlapping changes in surface gene. J Virol 88(12): 6805-6818.
- Kwon, S. Y., Park, Y. K., Ahn, S. H., Cho, E. S., Choe, W. H., Lee, C. H., Kim, B. K., Ko, S. Y., Choi, H. S., Park, E. S., Shin, G. C. and Kim, K. H. (2010). Identification and characterization of clevudine-resistant mutants of hepatitis B virus isolated from chronic hepatitis B patients. J Virol 84(9): 4494-4503.
- Kim, J. H., Park, Y. K., Park, E. S. and Kim, K. H. (2014). Molecular diagnosis and treatment of drug-resistant hepatitis B virus. World J Gastroenterol 20(19): 5708-5720.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ahn, S. H., Park, Y. K. and Kim, K. (2015). Introduction and Sequencing of Patient-isolated HBV RT Sequences into the HBV 1.2-mer Replicon. Bio-protocol 5(8): e1449. DOI: 10.21769/BioProtoc.1449.
Category
Microbiology > Microbial genetics > DNA > DNA replication
Microbiology > Antimicrobial assay > Antiviral assay
Microbiology > Microbe-host interactions > Virus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









