- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Real-time Analysis of Lateral Root Organogenesis in Arabidopsis
Published: Vol 5, Iss 8, Apr 20, 2015 DOI: 10.21769/BioProtoc.1446 Views: 10584
Reviewed by: Samik BhattacharyaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2323 Views

Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Pradeep Chand Deo
Oct 20, 2025 1438 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1691 Views
Abstract
Plants maintain capacity to form new organs such as leaves, flowers, lateral shoots and roots throughout their postembryonic lifetime. Lateral roots (LRs) originate from a few pericycle cells that acquire attributes of founder cells (FCs), undergo series of anticlinal divisions, and give rise to a few short initial cells. After initiation, coordinated cell division and differentiation occur, giving rise to lateral root primordia (LRP). Primordia continue to grow, emerge through the cortex and epidermal layers of the primary root, and finally a new apical meristem is established taking over the responsibility for growth of mature lateral roots [for detailed description of the individual stages of lateral root organogenesis see Malamy and Benfey (1997)]. To examine this highly dynamic developmental process and to investigate a role of various hormonal, genetic and environmental factors in the regulation of lateral root organogenesis, the real time imaging based analyses represent extremely powerful tools (Laskowski et al., 2008; De Smet et al., 2012; Marhavý et al., 2013; Marhavý et al., 2014). Herein, we describe a protocol for real time lateral root primordia (LRP) analysis, which enables the monitoring of an onset of the specific gene expression and subcellular protein localization during primordia organogenesis, as well as the evaluation of the impact of genetic and environmental perturbations on LRP organogenesis.
Materials and Reagents
- Arabidopsis seedlings (5-6 days old) expressing Green Fluorescent Protein (GFP) or analogous reporters (YFP, RFP, CFP, mCherry and others) (Chalfie et al., 1994; Shaner et al., 2007)
- MilliQ Water (H2O)
- Sucrose (VWR International, catalog number: 27483.294 )
- Murashige and Skoog basal salt mixture (MS salts) (Duchefa Biochemie, catalog number: M0221.0050 )
- 2- [N-morpholino] ethanesulfonic acid (MES) (Duchefa Biochemie, catalog number: M1503.0100 )
- Potassium hydroxide (KOH) (Merck KGaA, catalog number: 1.05021.1000 )
- Agar (LAB M, catalog number: MC029 )
- Ethanol (EtOH) (Sigma-Aldrich, catalog number: 32221 -2.5L)
- Seeds sterilization by ethanol (see Recipes)
- ½ MS media (see Recipes)
- Growth conditions (see Recipes)
Equipment
- Growth chamber to grow plant material
- Square plates 120 x 120 x 17 mm (Greiner Bio-One GmbH, catalog number: 688102 )
- Chambered cover glass (VWR, Kammerdeckgläser, Lab-TekTM, NuncTM - eine kammer, catalog number: 734-2056 )
- Inverted confocal microscope (Zeiss, model: LSM 700 )
Note: Fully motorized X, Y, Z scanning stage is required to perform multi-position time-lapse experiment.
- Objectives: 20x [suitable to monitor early phases of the lateral root primordia (LRP) initiation, Figure 2], 40x or 60x (suitable to monitor LRP from the stage I onwards, Figure 3)
- Fluorescence signal detection system for GFP and other fluorescent reporters (Shaner et al., 2007)
Software
- Software operating the confocal microscope
- ImageJ (Abramoff et al., 2004)
- CellseT (Pound et al., 2012)
- Microsoft Excel
Procedure
- Sample preparation (Figure 1)
- Prepare chambered cover glass, wash with 70% ethanol and dry (Figure 1A).
- Pour 45 ml MS+ medium into the square plate and wait till it congeals (approx. 40 min at room temperature; solid medium should be 2-3 mm thin) (Figure 1B).
- Using the chambered cover glass cut out the block of solid MS media (Figure 1C-D).
- From the block of media cut off a ~3 mm wide strip (Figure 1C-D).
- Using the strip of media grease the chambered cover glass (Figure 1E).
- Transfer 10-15 seedlings inside the chamber, roots of individual seedlings must not overlap (Figure 1F).
- Cover seedlings with remaining block of media (Figure 1G).
- Close with the chambered cover glass lid (Figure 1H).
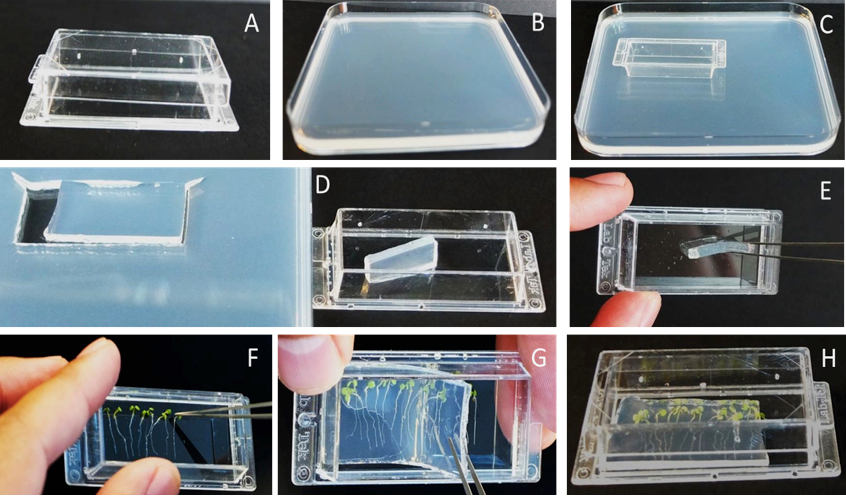
Figure 1. Sample preparation. A. Chambered cover glass. B. Solid MS+ medium 2 - 3 mm thin. C. Chambered cover glass used to cut the block of solid MS+ media. D. Small piece of MS+ media block is cut off. E. MS+ media block is used to grease the chambered cover glass. F. 10-15 Arabidopsis seedlings are transferred to the chamber. G. Arabidopsis seedlings are covered with remaining block of media. H. Closing with the chambered cover glass lid.
- Prepare chambered cover glass, wash with 70% ethanol and dry (Figure 1A).
- Real-time confocal imaging
- Prepare inverted confocal microscope for use [set lasers (for GFP-488); objectives - 40x/1.20 W; image size - x: 114.09 µm, y: 114.09 µm; zoom - 1.4; scan mode - plane, time series; pixel dwell - 1.27 µs; master gain - 652; digital gain - 1.5; digital offset - 0.00; pinhole 70 µm; filters - SP 555; beam splitters - MBS: MBS 405/488/555/639 DBS1: 492 nm].
Note: Parameters are to be adjusted according to specimen.
- Mount chamber with seedlings.
- Activate position list (list of marked positions of LRP).
- Find LRP at the stage of interest and focus at the middle plane of LRP (xylem pole strand adjacent to primordia must be in focus, Figures 2 and 3, time point 0). Mark position of the LRP. Move to next LRP and repeat the procedure. Optimal number of LRPs to be monitored is ~20 per experiment. To examine the process of founder cell (FC) specification and subsequent developmental phases we recommend to bend roots manually (Marhavy et al., 2013), to mark position of the root bent, focus on two xylem poles (Figure 2) and perform time-lapse imaging using objective 20x. To examine LRP development from stage I onwards we recommend performing time-lapse imaging using objective either 40x or 60x (dry, water or oil immersion).
- Activate time series.
- Set time intervals for scanning (typically 20 to 30 min). Keep in mind that with increasing number of LRPs over 30 you have to increase interval of scanning.
- Run time-lapse imaging. Typically, to follow LRP organogenesis from stage I till stage IV ~ 12 to 16 h observation time is needed. (In Arabidopsis thaliana LRP organogenesis involves eight developmental stages characterized by highly coordinated pattern of cell divisions and differentiation. Stage I: two pericycle founder cells divide asymmetrically to form primordia composed of up to ten short initial cells. Stage II: Initial cells divide periclinally forming an inner layer and an outer layer. Stages III and IV: The outer layer divides periclinally and the primordium consists of three layers (stage III) and later the inner layer undergoes a similar division, such that four cell layers are visible (stage IV). Stages V to VIII: Expansion and further division of the four layers eventually results in the emergence of the young lateral root from the parent tissue (the overlying tissue of the primary root) at stage eight. For details see Malamy and Benfey (1997).
- Process pictures for image analysis. Export confocal images in tif or jpg format; open images in ImageJ; proceed images to stack; and Save As an Avi format.
- Prepare inverted confocal microscope for use [set lasers (for GFP-488); objectives - 40x/1.20 W; image size - x: 114.09 µm, y: 114.09 µm; zoom - 1.4; scan mode - plane, time series; pixel dwell - 1.27 µs; master gain - 652; digital gain - 1.5; digital offset - 0.00; pinhole 70 µm; filters - SP 555; beam splitters - MBS: MBS 405/488/555/639 DBS1: 492 nm].
- Confocal imaging analysis
- To quantify the intensity of fluorescent reporter signal ImageJ might be used. Export and save the confocal pictures in TIF format to analyze data using ImageJ. Or, alternatively, import the confocal stacks into imageJ by the BioFormats plugin. Open image in ImageJ; using segmented line (width of the line adjusted accordingly); mark the area of interest and use function “Mean” to calculate average intensity in pixels. Copy the results “Mean” to Excel program for further processing.
- To determine polar localization of fluorescently labeled membrane proteins CellseT software might be used (Pound et al., 2012). The software is suitable to evaluate cell and tissue geometry as well as to quantify the intensity of fluorescent marker signal. Follow CellseT instructions for further details.
- To evaluate dynamics of LRP development time lapse series images might be processed into AVI file (Windows media player or comparable program).
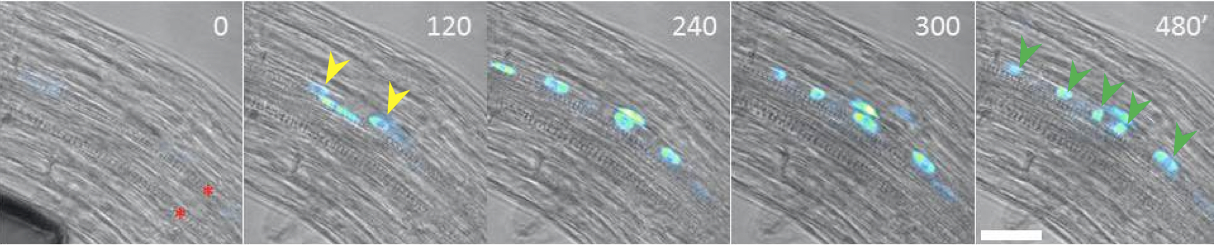
Figure 2. Real-time analysis of founder cell establishment and early phases of LRP initiation using DR5pro::N7:Venus auxin reporter (Heisler et al., 2005). Accumulation of the nuclear DR5pro::N7:Venus signal in two pericycle cells at 120 min. indicates establishment of FCs (yellow arrows). Anticlinal divisions occurring between 240-480 min give rise LRP at developmental stage A composed of 5 initial cells (green arrows). To observe FC establishment root were manually bent prior monitoring. Objective 20x used for observation. Time in minutes (upper right corner) is relative to root bending. Red asterisks indicate two xylem poles. Scale bar: 40 μm
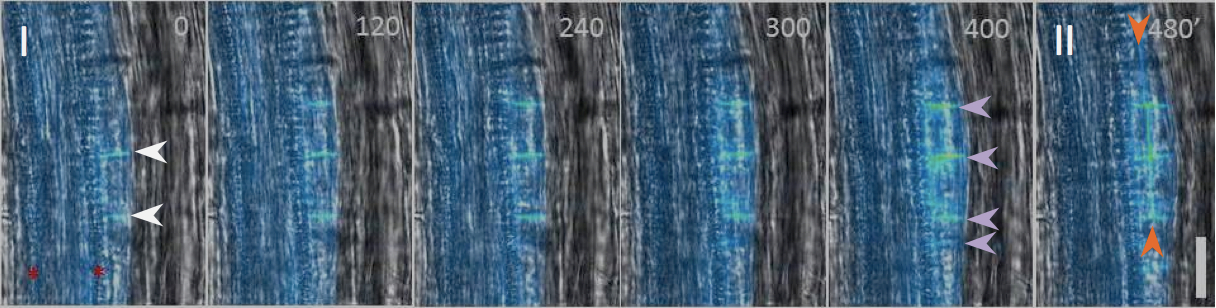
Figure 3. Real-time analysis of the LRP development using auxin efflux carrier PIN1::PIN1-GFP reporter (Benkova et al., 2003). PIN1::PIN1-GFP localizes to cell membranes (white arrows) and is expressed from LRP stage I onwards. In time interval 400-480 min LRP which composes of 5 initial cells (pink arrows) undergoes periclinal division (orange arrows) and transits to developmental stage II. Time in minutes (upper right corner). Red asterisks mark two xylem poles. Scale bar: 30 μm
- To quantify the intensity of fluorescent reporter signal ImageJ might be used. Export and save the confocal pictures in TIF format to analyze data using ImageJ. Or, alternatively, import the confocal stacks into imageJ by the BioFormats plugin. Open image in ImageJ; using segmented line (width of the line adjusted accordingly); mark the area of interest and use function “Mean” to calculate average intensity in pixels. Copy the results “Mean” to Excel program for further processing.
Recipes
- Seeds sterilization by ethanol
- Transfer seeds in 2 ml Eppendorf tubes (maximum volume of seeds to be sterilized per tube should not exceed 3 mm from bottom).
- Add 1 ml 70% EtOH (technical grade is sufficient), shake for 5 sec and leave seeds to sediment for 10 min.
- Remove 70% EtOH.
- Wash seeds in 100% EtOH under the clean bench.
- Dry seeds under the clean bench.
- Transfer seeds in 2 ml Eppendorf tubes (maximum volume of seeds to be sterilized per tube should not exceed 3 mm from bottom).
- ½ MS media (1 L)
- Add 10 g sucrose
- Add 2.3 g MS Salts
- Add 0.5 g MES
- Adjust pH to 5.9 (KOH)
- Add 8 g agar (1,000 ml bottle)
- H2O
- Add 10 g sucrose
- Growth conditions
- Seeds of Arabidopsis were plated on square plates filled with MS+ medium (45.5 ml).
- Stratification for 2 days at 4 °C in dark.
- Seedlings were grown on vertically oriented plates in growth chambers under a 16-h-light/8-h-dark photoperiod at 18 or 21 °C.
- Seeds of Arabidopsis were plated on square plates filled with MS+ medium (45.5 ml).
Acknowledgments
We thank Matyas Fendrych for critical reading and comments. This work was supported by the European Research Council with a Starting Independent Research grant (ERC-2007-Stg-207362-HCPO) and the Czech Science Foundation (GA13-39982S) to Eva Benková. The protocol was developed based on previously published work of De Rybel et al. (2010) and Laskowski et al. (2008).
References
- Abràmoff, M. D., Magalhães, P. J. and Ram, S. J. (2004). Image processing with ImageJ. Biophotonics Int 11(7): 36-43.
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G. and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115(5): 591-602.
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. and Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science 263(5148): 802-805.
- De Smet, I., White, P. J., Bengough, A. G., Dupuy, L., Parizot, B., Casimiro, I., Heidstra, R., Laskowski, M., Lepetit, M., Hochholdinger, F., Draye, X., Zhang, H., Broadley, M. R., Peret, B., Hammond, J. P., Fukaki, H., Mooney, S., Lynch, J. P., Nacry, P., Schurr, U., Laplaze, L., Benfey, P., Beeckman, T. and Bennett, M. (2012). Analyzing lateral root development: how to move forward. Plant Cell 24(1): 15-20.
- De Rybel, B., Vassileva, V., Parizot, B., Demeulenaere, M., Grunewald, W., Audenaert, D., Van Campenhout, J., Overvoorde, P., Jansen, L., Vanneste, S., Moller, B., Wilson, M., Holman, T., Van Isterdael, G., Brunoud, G., Vuylsteke, M., Vernoux, T., De Veylder, L., Inze, D., Weijers, D., Bennett, M. J. and Beeckman, T. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20(19): 1697-1706.
- Dubrovsky, J. G. and Forde, B. G. (2012). Quantitative analysis of lateral root development: pitfalls and how to avoid them. Plant Cell 24(1): 4-14.
- Laskowski, M., Grieneisen, V. A., Hofhuis, H., Hove, C. A., Hogeweg, P., Maree, A. F. and Scheres, B. (2008). Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6(12): e307.
- Malamy, J. E. and Benfey, P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124(1): 33-44.
- Marhavy, P., Bielach, A., Abas, L., Abuzeineh, A., Duclercq, J., Tanaka, H., Parezova, M., Petrasek, J., Friml, J., Kleine-Vehn, J. and Benkova, E. (2011). Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21(4): 796-804.
- Marhavý, P., Vanstraelen, M., De Rybel, B., Zhaojun, D., Bennett, M. J., Beeckman, T. and Benková, E. (2013). Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J 32(1): 149-158.
- Marhavý, P., Duclercq, J., Weller, B., Feraru, E., Bielach, A., Offringa, R., Friml, J., Schwechheimer, C., Murphy, A. and Benkova, E. (2014). Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr Biol 24(9): 1031-1037.
- Shaner, N. C., Patterson, G. H. and Davidson, M. W. (2007). Advances in fluorescent protein technology. J Cell Sci 120(Pt 24): 4247-4260.
- Pound, M. P., French, A. P., Wells, D. M., Bennett, M. J. and Pridmore, T. P. (2012). CellSeT: novel software to extract and analyze structured networks of plant cells from confocal images. Plant Cell 24(4): 1353-1361.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Marhavý, P. and Benková, E. (2015). Real-time Analysis of Lateral Root Organogenesis in Arabidopsis. Bio-protocol 5(8): e1446. DOI: 10.21769/BioProtoc.1446.
Category
Plant Science > Plant cell biology > Cell structure
Plant Science > Plant developmental biology > Morphogenesis
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









