- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of 70S Ribosomes from Bacillus subtilis
Published: Vol 5, Iss 7, Apr 5, 2015 DOI: 10.21769/BioProtoc.1432 Views: 12430
Reviewed by: Elizabeth LibbyManuela RoggianiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Lauren E.-A. Eyssen [...] Raymond J. Owens
Mar 20, 2024 6244 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2096 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views
Abstract
The eubacterial ribosome (70S) is a macromolecular complex that is composed of a small (30S) subunit and a large (50S) subunit. The small subunit comprises the 16S ribosomal RNA (rRNA) and more than 20 ribosomal proteins (r-proteins), whereas the large subunit comprises the 23S and 5S rRNAs and more than 30 r-proteins. Bacillus subtilis (B. subtilis) has 57 r-ribosomal protein genes and three rRNAs (16S, 23S and 5S rRNAs). Among them, we identified 21 r-proteins of the small subunit and 31 r-proteins of the large subunit in B. subtilis (Nanamiya et al., 2004). The functions and roles of individual components of the ribosome have not yet been completely clarified. Herein we describe in detail an ultracentrifugation-based protocol for the preparation of 70S ribosomes from exponentially growing cells of B. subtilis.
Keywords: Bacillus subtilisMaterials and Reagents
- Bacillus subtilis 168
- NaCl
- BD BactoTM tryptone (Difco, catalog number: 211705 )
- BD BactoTM yeast extract (Difco, catalog number: 212750 )
- Agar (Nissui, catalog number: 05835 )
- Tris (Wako Pure Chemical Industries, catalog number: 512-97505 )
- Magnesium acetate (Wako Pure Chemical Industries, catalog number: 139-15335 )
- Ammonium acetate (Wako Pure Chemical Industries, catalog number: 019-02835 )
- Dithiothreitol (DTT) (Wako Pure Chemical Industries, catalog number: 042-29222 )
- Phenylmethylsulphonyl fluoride (PMSF)
- Sucrose (Sigma-Aldrich, catalog number: S0389-55G )
- Diethylpyrocarbonate (DEPC) treated water
Note: DEPC was added to distilled water at a final concentration of 1%, and then it stored at room temperature for over two hours. The DEPC-treated distilled water was autoclaved twice (121 °C, 20 min) and stored at room temperature. - LB medium (see Recipes)
- Buffer I (see Recipes)
- 10-40% sucrose solutions (see Recipes)
Equipment
- 2 L flasks with cotton plugs
- Innova 4080 bench top incubator shaker (New Brunswick scientific)
- French pressure cell press [Aminco International (USA), model: FA-078 ]
- Mini cell [Aminco International (USA), model: FA-003 ]
- Gradient fractionator (BIOCOMP, catalog number: 152-001 )
- Gradient master [Aminco International (USA), catalog number: 107-201M ]
- Micro collector (ATTO Technology, catalog number: AC-5700P )
- Nano drop 2000 (Thermo Fisher Scientific)
- High-speed refrigerated centrifuge (e.g. Hitachi, model: himac CR22GII )
- Angle rotor R10A3 (fixed angle, max: 10,000 rpm,18,800 x g, volume: 500 ml x 6) (Hitachi)
- Centrifuge MX-300 (TOMY)
- TMA-300 rotor (fixed angle, max: 15,000 rpm, 20,380 x g, volume: 2 ml x 24) (TOMY)
- TMA-27 rotor (fixed angle, max: 15,000 rpm, 21,130 x g, volume: 15 ml x 4, 50 ml x 4) (TOMY)
- Centrifuge tube (Nalgene, catalog number: 3148-0050 )
- Ultra-centrifuge (e.g. Hitachi, model: CP-60E )
- P55ST2 rotor (swinging bucket rotor, max: 55,000 rpm, 366,000 x g, volume: 5 ml x 6) (Hitachi)
- P28S rotor (swinging bucket rotor, max: 28,000 rpm, 141,000 x g, volume: 40 ml x 6) (Hitachi)
- P40ST rotor (swinging bucket rotor, max: 40,000 rpm, 284,000 x g, volume: 13 ml x 6) (Hitachi)
- Open-top polyclear centrifuge tube for P40ST rotor (Seton Identification Products, part number: 7031 )
- Open-top polyclear centrifuge tube for P28S rotor (Seton Identification Products, part number: 7052 )
- Centrifuge ware for P55ST2 rotor (Hitachi, part number: 332245A )
Note: These rotors can substitute for other commercial rotors which can be used by the same centrifugal force as described in procedures.
Procedure
- Crude preparation of 70S ribosomes
- B. subtilis is streaked out for single colonies and pre-incubated on LB agar plates at 30 °C for 14-16 h.
- Cells are taken from single colonies on the plates and suspended in approximately 10 ml of LB medium with high turbidity. The cell suspension was diluted into 3 L of LB medium at an OD600 of 0.04, and distributed equally among six 2-liter flasks, each containing 500 ml of the cell culture. The flasks are incubated at 37 °C with shaking at 250 rpm.
Note: B. subtilis is routinely cultured in volumes that do not exceed one quarter of the volume of the flask used. - Cells are harvested by centrifugation, when the OD600 of the culture reaches 0.2, using six 500-ml centrifuge tubes (7,000 x g; 5 min, 4 °C). Cell pellets are resuspended with small volume of the culture supernatant, transferred from a 500-ml centrifugation tube to six 15-ml sample tubes and pellets are collected (3,500 x g, 5 min, 4 °C). The cell pellets are frozen by liquid N2.
- Each cell pellet is suspended in 3.5 ml of Buffer I pre-chilled on ice.
- Cells are disrupted using a French press (8,000 p.s.i.) passing the sample through three times.
- Cell debris is removed by centrifugation (11,000 x g, 30 min, 4 °C).
- The supernatant (cell lysate) is centrifuged at 30,000 x g for 30 min at 4 °C, for example in a HITACHI P55ST2 rotor at 18,000 rpm.
- The resulting supernatant (S30 supernatant) is centrifuged at 200,000 x g for 100 min at 4 °C (e.g. in a HITACHI P55ST2 rotor at 45,000 rpm).
- Each pellet is resuspended with 200 μl of Buffer I and the suspension stored at -80 °C until required (S100 pellet fraction).
Note that the S100 fraction can be seen as amber pellet at bottom of the tube. Using a glass rod may be preferable to resuspend the pellet because resuspension of the pellet by pipetting is not easy.
Note: Care is taken to avoid generating foam while resuspending the pellet.
- B. subtilis is streaked out for single colonies and pre-incubated on LB agar plates at 30 °C for 14-16 h.
- Purification of 70S ribosomes
- To further purify the 70S ribosomes, prepare a 10-40% sucrose gradient in SW28 centrifuge tubes using a Gradient Master gradient mixer.
- 10% and 40% sucrose solutions are prepared in Buffer I.
- The 10 to 40% sucrose gradient is generated in a SW28 centrifuge tube using a Gradient Master gradient mixer.
- The S100 pellet fraction is applied on the top of the 10 to 40% sucrose gradient and centrifuged at 67,000 x g for 17 h at 4 °C (e.g. in a HITACHI P28S rotor at 22,500 rpm, using 270 A260 units per centrifuge tube).
- Samples are taken by Piston Gradient Fractionator and Micro collector. Absorbance profiles at 260 nm are determined using a Nano drop 2000.
Note: An example of the ribosome profile from crude cell extracts is shown in Figure 1. - Collected fractions containing 70S ribosomes are identified as a peak with an absorbance at 260 nm.
- The ribosomes are recovered from the pooled fractions, which are diluted with Buffer I (1:2 dilutions) and centrifuged at 64,000 x g for 17 h at 4 °C. (e.g. in a HITACHI P40ST rotor at 22,500 rpm).
- The supernatant is removed completely from the centrifuge tubes.
- The resulting 70S ribosome pellets are covered with 300 μl of Buffer I and left overnight at 4 °C. The pellet is then gently resuspended in Buffer I using a glass rod and stored at -80 °C until required.
Note: Care is taken to avoid generating foam while resuspending the pellet.
- To further purify the 70S ribosomes, prepare a 10-40% sucrose gradient in SW28 centrifuge tubes using a Gradient Master gradient mixer.
Representative data
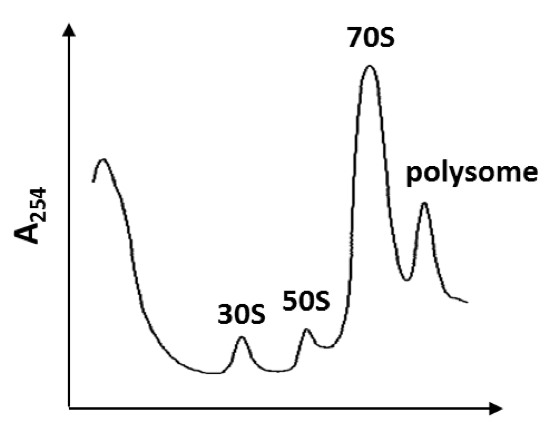
Figure 1. Profile of ribosome from crude cell extract by 10-40% sucrose gradient sedimentation. The crude cell extract prepared from the log phase cells (OD600 = 0.2) of wild type strain (168 trpC2) was applied on the top of the 10 to 40% sucrose gradient and centrifuged at 55,000 x g for 17.5 h at 4 °C (e.g. in a HITACHI P40ST rotor at 21,000 rpm, using 10 A260 units per centrifuge tube). The A260 measurement shows as the same profile pattern of A420 measurement, and the peak of 70S ribosome can be determined. When the S100 fraction was subjected to the 10 to 40% sucrose gradient and centrifugation, the peak smaller than 30S was not observed.
Recipes
- LB medium
0.5% NaCl
1% tryptone
0.5% yeast extract
1.5% agar (only use for plates) - Buffer I (200 ml is enough, pre-chilled on ice)Buffer I is prepared with DEPC treated water.
20 mM Tris-HCl (pH 7.6) 15 mM magnesium acetate 100 mM ammonium acetate 0.1 mM DTT 2 mM PMSF
Note: Buffer I can be stored without DTT and PMSF. DTT and PMSF are added to the buffer before use. Buffer I should be pre-chilled throughout the entire procedure. - 10-40% sucrose solutions
10-40% sucrose solutions are made in Buffer I.
Acknowledgments
This protocol was adapted from Nanamiya et al. (2004) and Suzuki et al. (2014). This work was supported in part by Grants-in-Aid for Scientific Research (C) (G. A.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Nanamiya, H., Akanuma, G., Natori, Y., Murayama, R., Kosono, S., Kudo, T., Kobayashi, K., Ogasawara, N., Park, S. M., Ochi, K. and Kawamura, F. (2004). Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol Microbiol 52(1): 273-283.
- Nanamiya, H., Kawamura, F. and Kosono, S. (2006). Proteomic study of the Bacillus subtilis ribosome: Finding of zinc-dependent replacement for ribosomal protein L31 paralogues. J Gen Appl Microbiol 52(5): 249-258.
- Natori, Y., Nanamiya, H., Akanuma, G., Kosono, S., Kudo, T., Ochi, K. and Kawamura, F. (2007). A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol 63(1): 294-307.
- Suzuki, S., Tanigawa, O., Akanuma, G., Nanamiya, H., Kawamura, F., Tagami, K., Nomura, N., Kawabata, T. and Sekine, Y. (2014). Enhanced expression of Bacillus subtilis yaaA can restore both the growth and the sporulation defects caused by mutation of rplB, encoding ribosomal protein L2. Microbiology 160(Pt 6): 1040-1053.
- Tagami, K., Nanamiya, H., Kazo, Y., Maehashi, M., Suzuki, S., Namba, E., Hoshiya, M., Hanai, R., Tozawa, Y., Morimoto, T., Ogasawara, N., Kageyama, Y., Ara, K., Ozaki, K., Yoshida, M., Kuroiwa, H., Kuroiwa, T., Ohashi, Y. and Kawamura, F. (2012). Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. Microbiologyopen 1(2): 115-134.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Suzuki, S., Akanuma, G. and Kawamura, F. (2015). Purification of 70S Ribosomes from Bacillus subtilis. Bio-protocol 5(7): e1432. DOI: 10.21769/BioProtoc.1432.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Microbiology > Microbial biochemistry > RNA
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









