- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Substrate Specificity of Recombinant Ser/Thr Protein Kinase
Published: Vol 5, Iss 6, Mar 20, 2015 DOI: 10.21769/BioProtoc.1426 Views: 9948
Reviewed by: Maria SinetovaYoko EguchiTatsuki Kunoh

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies
Parthasarathy Manikandan [...] Mahavir Singh
Jul 5, 2023 2174 Views

Isolation of Antigen-Specific Nanobodies From Synthetic Libraries Using a Protein Selection Strategy That Combines MACS-Based Screening of YSD and FLI-TRAP
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
Jan 20, 2026 474 Views
Abstract
Protein kinases are enzymes that phosphorylate proteins in a cell. Determination of kinase activity in reactions of phosphorylation is a very convenient way for a biochemical characterization of this group of enzymes. Here we describe a method to determine the activity of a recombinant Ser/Thr protein kinase using as a possible substrate MBP, H1, and BSA.
Materials and Reagents
- Escherichia coli (E.coli) strains - for cloning and for expression - DH5α, Rosetta (DE3 pLysS)
- Specific synthetic oligonucleotides
- PCR reagents (dNTP, MgCl2, PCR buffer, Taq-polymerase, DNA) (Thermo Fisher Scientific, catalog numbers: R0181 , EP0612 )
- Plasmids for cloning and for expression [pTZ57R/T, pET41a(+)]
- CaCl2 (Sigma-Aldrich, catalog number: C3306-250G ) (for transformation)
- Antibiotics (Ampicillin, Kanamycine, Chloramphenicol) (AppliChem, catalog numbers: A-0839 , A-1493 , A-6435 )
- Isopropyl thio-β-D-galactoside (IPTG) (Fermentas, catalog number: R0393 )
- Tris-HCl (Sigma-Aldrich, catalog number: T6791 )
- Ethylenediaminetetraacetic acid (EDTA) (AppliChem, catalog number: A-1104.0500 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815-5G )
- Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889-10G )
- Na3VO4 (Sigma-Aldrich, catalog number: S6508-10G )
- NaF (Sigma-Aldrich, catalog number: S7920-100G )
- β-glycerophosphate (Sigma-Aldrich, catalog number: G9422-50G )
- Sucrose (AppliChem, catalog number: A-1125 )
- Phenylmethylsulfonyl fluoride (PMSF) (AppliChem, catalog number: A-0999 )
- Benzamidine hydrochloride (Sigma-Aldrich, catalog number: B6506 )
- 6-aminocaproic acid (Sigma-Aldrich, catalog number: A7824-25G )
- 2-(N-morpholino)ethanesulfonic acid (MES) (Amresco, catalog number: Am-E169-0.05 )
- Myelin Basic Protein bovine (MBP) (Sigma-Aldrich, catalog number: M1891-5MG )
- Histone from calf thymus (H1) (Sigma-Aldrich, catalog number: H5505-100MG )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: 05470-1G )
- MgCl2 (AppliChem, catalog number: A-1447 )
- Brilliant Blue R Concentrate (Sigma-Aldrich, catalog number: B8647 )
- BCA Protein Assay Kit (Pierce Thermo Scientific, catalog number: 23227 )
- Sodium dodecyl sulfate, SDS (Sigma-Aldrich, catalog number: L3771 )
- TGX FastCast Acrylamide Starter Kit (12%) (Bio-Rad Laboratories, catalog number: 161-0175 )
- N,N,N',N'-tetramethylethylenediamine (TEMED) (Bio-Rad Laboratories, catalog number: 161-0800 )
- Ammonium Persulfate (APS) (Bio-Rad Laboratories, catalog number: 161-0700 )
- Illustra NAP-5 Columns (GE Healthcare Life Sciences, catalog number: 17-0853-02 )
- [γ-32P] ATP
- Homogenization buffer (see Recipes)
- Reaction mixture (see Recipes)
Equipment
- Temperature controlled shaker (Heidolph, model: Unimax 1010 )
- French Pressure Cell Press (Aminco) or glass beads (Sigma-Aldrich, catalog number: G4649 ) for cell disruption
- Thermostat
- Spectrophotometer
- Gel electrophoresis equipment (Mini-PROTEAN Tetra Handcast Systems, Bio-Rad Laboratories, catalog number: 165-8000 )
- Laminar flow box
- Refrigerated microcentrifuge preset to 4 °C (Eppendorf, model: 5415 R )
- Gel Dryer System (Bio-Rad Laboratories, catalog number: 165-1789 )
- Exposure cassette/BioMax® intensifying screen set (Sigma-Aldrich, catalog numbers: C4979 and P8432 )
- Carestream® Kodak® autoradiography GBX developer/replenisher (Sigma-Aldrich, catalog number: P7042 )
- Carestream® Kodak® autoradiography GBX fixer/replenisher (Sigma-Aldrich, catalog number: P7167 )
Procedure
- Expression and isolation of a recombinant protein kinase
- Amplify a DNA fragment carrying the gene of interest using synthetic primers. For the more convenient cloning of the desired insert, it is worth to construct the primers harboring specific restriction sites for endonucleases. The easiest way to clone this PCR product is to use T-vector pTZ57R/T. Transform DH5-α strain. Spread transformed cell onto LB agar plates containing ampicillin, IPTG, X-GAL. Incubate overnight at 37 °C. Select white colonies that should harbor plasmid with a desired insert and inoculate 3 ml of LB-medium. Extract pDNA from the overnight culture. Check it for containing your full length insert either via PCR or via restriction analysis.
- Ligate your restriction fragment with the appropriate expression vector.
- Transform E. coli expression strain with the resultant plasmid using CaCl2 method (Sambrook et al., 1989). Spread transformed cells on the LB-plate containing specific antibiotic that is coded by the expression plasmid. Manipulations with bacterial cells should be carried out in a flow box (He, 2011).
- Grow the transformed cells in 100 ml of LB-medium until the OD595 of suspension reached 0.5. Initiate overproduction of the recombinant protein by the addition of IPTG to a final concentration of 0.1 mM. Induction conditions may vary strongly (from 3 h at 37 °C to 15 h at 25 °C). Centrifuge cells at 3,500 x g for 10 min at 4 °C.
- Resuspend the collected cells in a prechilled homogenization buffer (1 g of cells in up to 5 ml of buffer). Disrupt the cells either using a French Pressure Cell Press (2.8 MPa) or glass beads (use an equal volume of beads and cell suspension. Vortex vigorously for 30 sec. To avoid warming of the sample incubate a test tube for 30 sec on ice. Repeat this procedure 10 times). All procedures should be carried out at 4 °C.
- Centrifuge the crude extracts stepwise, first at 7,500 x g for 15 min to remove unbroken cells and afterwards at 16,000 x g for 20 min for clarifying cell lysate. Procedures should be carried out at 4 °C. Collect supernatant (soluble protein fraction) and keep a pellet.
- Desalt the soluble protein fraction on NAP-5 columns according manufacturer’s instruction. Elute the protein with 20 mM MES-КОН (pH 6.5) and determine protein concentration using BCA Protein Assay Kit. To determination protein quantity in the pellet resuspend it with 4% of SDS and recover the proteins by centrifugation at 16,000 x g for 20 min. Then determine protein concentration with BCA Protein Assay Kit. Mix protein sample with appropriate amount of Laemmli sample buffer to get the final protein concentration 1 mg/ml (He, 2011).
- Prepare and run SDS-PAGE. Stain the gel with CBB R250 (Yan, 2011).
- Analyze both the desalted soluble fraction and proteins from the pellet using SDS-PAGE. If your protein is soluble proceed to the protein kinase assay. If the protein is in the pellet, it is necessary to recover the protein from inclusion bodies or to start over induction procedure in other conditions, trying to shift the protein into the supernatant. Alternatively, purify the soluble proteins further by affinity chromatography. The first possibility allows to obtain the result much quickly. The second one requires more time and an additional purification step but gives the better picture with the fewer non specific bands.
- Amplify a DNA fragment carrying the gene of interest using synthetic primers. For the more convenient cloning of the desired insert, it is worth to construct the primers harboring specific restriction sites for endonucleases. The easiest way to clone this PCR product is to use T-vector pTZ57R/T. Transform DH5-α strain. Spread transformed cell onto LB agar plates containing ampicillin, IPTG, X-GAL. Incubate overnight at 37 °C. Select white colonies that should harbor plasmid with a desired insert and inoculate 3 ml of LB-medium. Extract pDNA from the overnight culture. Check it for containing your full length insert either via PCR or via restriction analysis.
- Phosphorylation in vitro
- Kinase activity assay is performed in a reaction mixture containing 20 mM MES-КОН (pH 6.5), 10 mM MgCl2, 10 µM АТP, exogenous protein substrate, 74 kBq 2μCi [γ-32P] ATP (specific activity 148 TBq/mmol). The quantity of the labeled ATP depends on the quantity of the protein. Different proteins (histone Н1, basic myelin protein МВР, and BSA) can be used as possible exogenous substrates. To calculate the volume of the [γ-32P] ATP required you can use, for example, this site http://www.radprocalculator.com/Decay.aspx. Or you can do it manually:
Radioactive concentration (µCi/µl) =Calibration Concentration (µCi)/2 N/14.3,
where N - number of days passed from the day of calibration, taken from the passport of the product. 14.3 days - half-life period of the 32P.
The reaction is initiated by the addition of 5-100 µg of the recombinant protein kinase and allowed to proceed for 15 min at 30 °C. The quantity of the recombinant protein kinase in this reaction depends on whether affinity purified or non-purified the recombinant protein kinase is used.
- The reaction is terminated by adding half of the reaction volume of 3x Laemmli sample buffer and samples are boiled for 5 min.
- Phosphorylated products are resolved on SDS-polyacrylamide gels. The gels are then stained with CBB R250 as mentioned above, dried, and subjected to autoradiography. Exposition of dried gels with the X-Ray film varies from few hours at room temperature to few days at -70 °C and depends on the quantity and the activity of the recombinant protein kinase.
- If the recombinant protein kinase is enzymatically active it can demonstrate either autophoshorylation or phosphorylation of the substrate exogenously provided. So, it is possible to make a decision about substrate specificity of the recombinant protein kinase examined. According to the results obtained we can conclude that at the phosphorylation condition described here the examined recombinant protein kinase is capable to phosphorylate histone H1. Since at the autoradioradiograph there is no a signal corresponding to apparent molecular weight of the recombinant protein kinase it does not undergo autophosphorylation.
- Kinase activity assay is performed in a reaction mixture containing 20 mM MES-КОН (pH 6.5), 10 mM MgCl2, 10 µM АТP, exogenous protein substrate, 74 kBq 2μCi [γ-32P] ATP (specific activity 148 TBq/mmol). The quantity of the labeled ATP depends on the quantity of the protein. Different proteins (histone Н1, basic myelin protein МВР, and BSA) can be used as possible exogenous substrates. To calculate the volume of the [γ-32P] ATP required you can use, for example, this site http://www.radprocalculator.com/Decay.aspx. Or you can do it manually:
Representative data
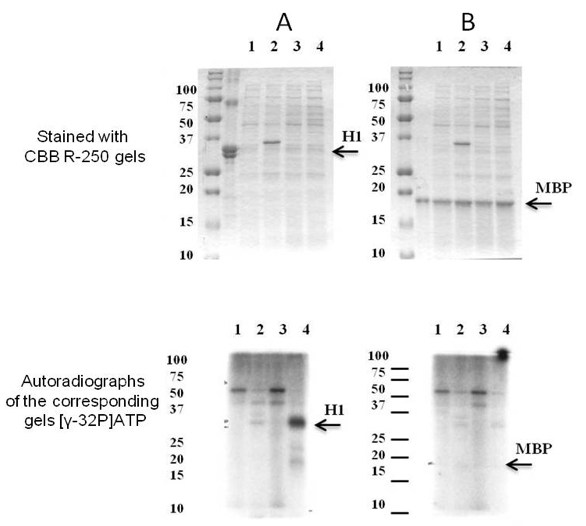
Figure 1. The substrate specificity of recombinant SpkE. Coomassie R-250 staining (at the top) and autoradiographs of the corresponding gels (at the bottom). As potential substrates were examined (A) histone H1 and (B) MBP. Soluble proteins were isolated from E. coli cells with pET41a(+) (1) before and (2) after induction with IPTG and E. coli cells with pET41a(+) carrying Synechocystis spkE (3) before and (4) after induction with IPTG. About 10 μg of the E. coli cells soluble protein fraction was mixed in 25 μl with 5 μg either H1 or MBP in the presence of 1.5 mCi of [γ-32P]ATP. The phosphorylation reaction was carried out for 15 min at 30 °C. As demonstrated above only histone H1 is phosphorylated by SpkE. [modified from Zorina et al. (2014)]
Recipes
- Homogenization buffer
50 mM Tris–HCl (pH 7.6)
10 mM MgCl2
2 mM EDTA
1 mM DTT
1 mM EGTA
2 mM Na3VO4
10 mM NaF
50 mM β-glycerophosphate
250 mM sucrose
1 mM PMSF
1 mM benzamidine hydrochloride
0.5 mM 6-aminocaproic acid
- Reaction mixture (total volume 25 µl)
20 mM MES-KOH (pH 6.5)
10 mM MgCl2
10 µM ATP (unlabelled)
0.25 mg/ml MBP, or 0.5 mg/ml H1, or 1 mg/ml BSA
2 µCi [γ-32P]ATP
Protein kinase
ddH2O
Acknowledgments
This work was supported by the grant from Russian Science Foundation no. 14-24-00020.
References
- He, F. (2011). Standard DNA cloning. Bio-protocol Bio101: e52.
- He, F. (2011). Laemmli-SDS-PAGE. Bio-protocol Bio101: e80.
- Sambrook, J., Fritsch, E. F. and Maniatis, T. (1989). Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press.
- Yan, Q. (2011). Rapid coomassie protein SDS-gel staining. Bio-protocol Bio101: e83.
- Zorina, A. A., Bedbenov, V. S., Novikova, G. V., Panichkin, V. B. and Los, D. A. (2014) Involvement of serine/threonine protein kinases in the cold stress response in the cyanobacterium Synechocystis sp. PCC 6803: Functional characterization of SpkE protein kinase. Mol Biol (Moscow) 48: 390–398.
- Zorina, A., Stepanchenko, N., Novikova, G. V., Sinetova, M., Panichkin, V. B., Moshkov, I. E., Zinchenko, V. V., Shestakov, S. V., Suzuki, I., Murata, N. and Los, D. A. (2011). Eukaryotic-like Ser/Thr protein kinases SpkC/F/K are involved in phosphorylation of GroES in the Cyanobacterium synechocystis. DNA Res 18(3): 137-151.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zorina, A. A., Novikova, G. V. and Los, D. A. (2015). Substrate Specificity of Recombinant Ser/Thr Protein Kinase. Bio-protocol 5(6): e1426. DOI: 10.21769/BioProtoc.1426.
Category
Microbiology > Microbial biochemistry > Protein > Interaction
Biochemistry > Protein > Interaction > Protein-protein interaction
Cell Biology > Cell signaling > Phosphorylation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










