- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Chemiluminescence Detection of the Oxidative Burst in Plant Leaf Pieces
Published: Vol 5, Iss 6, Mar 20, 2015 DOI: 10.21769/BioProtoc.1423 Views: 19160
Reviewed by: Tie LiuMasahiro Morita

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rice Ragged Stunt Virus Propagation and Infection on Rice Plants
Chao Zhang [...] Jianguo Wu
Oct 20, 2018 6505 Views

Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries
Piao Yang [...] Ye Xia
Oct 20, 2023 2776 Views
Abstract
The production of 'reactive oxygen species' (ROS), also termed oxidative burst, is a typical cellular response of animals and plants to diverse biotic and abiotic stresses. Here, we outline the detection of the ROS-burst in plant leaf pieces using a luminol-based bioassay which allows for the detection of chemiluminescence. The assay was originally described by Keppler et al. (1989) and subsequently adapted for other plant cells and tissues (Felix et al., 1999) and also used in recent publications (Albert et al., 2013; Albert et al., 2010; Butenko et al., 2014; Halter et al., 2014). In this protocol we outline a standardized version of this assay including remarks and recommendations for data evaluation and interpretation of results.
Materials and Reagents
- Plant leaf pieces (any plant of interest, e.g. Arabidopsis or N. benthamiana transiently expressing a construct of interest)
- Water
- 200 µM luminol L-012 (Wako Chemicals USA, catalog number: 075-05111 ; http://www.wako-chem.co.jp/english/labchem/journals/wpu_bio1/10.htm)
- 10 µg/ml peroxidase, horseradish peroxidase (AppliChem, catalog number: A3791 )
- MAMPs or any elicitors of interest
- Known MAMP as positive control (e.g. flg22, chitin-oligomers, etc.)
- 10x luminol master mix (see Recipes)
Equipment
- Scissors
- Petridishes
- Small spatula
- 96 well plates, white, flat-bottomed (e.g., LIA-plate, whit, 96-well, flat-bottom, Greiner Bio-One GmbH) or suitable cuvettes for single cell instruments
- Luminometer, either a 96 well plate reader (e.g., Centro LB 960, Microplate Luminometer, BERTHOLD TECHNOLOGIES) or a single cell instrument
Procedure
- Preparation of plant samples, one day before oxidative burst measurements
- Label petridishes appropriately and fill half with deionized water.
- Cut leaves (younger or highly differentiated) of plants [important: plants should not flower, and should not be too old (about 4-6 weeks)] with scissors and cut off the edge of the leaves with scissors.
- Cut the remaining part of the leaf in small, equally sized pieces (e.g. 3 x 3 mm, pieces should easily fit into the wells of a 96-well plate). It is recommended that you cut about 2x to 3x more leaf pieces than you need.
- Float leaf pieces on the water in the petridishes (orientation does not matter), avoid submersion!
- Cover the samples and incubate overnight (or > 8 h) at room temperature, avoid shaking.
- Label petridishes appropriately and fill half with deionized water.
- Preparation of samples prior to luminescence measurements, next day
- Depending on the luminometer (single cell or multi-well plate reader) 90 µl water + 10 µl luminol master mix (10x) are pipetted into each well of a new 96-well plate. For single cell luminometers this might be scaled up 2x to 3x, according to the size of the measurement cuvette.
- Carefully add one leaf piece per well, avoid mechanical damage and wounding! If the pieces are very small one could also use two per well. Use a small spatula, no forceps! Check if all leaf pieces are in the center of the well prior to starting the measurement.
- Per planned treatment prepare about 3-4 replicates (each in an extra well); do not forget negative (e.g. untreated) and positive controls; all leaf pieces should have the equal size!
- Depending on the luminometer (single cell or multi-well plate reader) 90 µl water + 10 µl luminol master mix (10x) are pipetted into each well of a new 96-well plate. For single cell luminometers this might be scaled up 2x to 3x, according to the size of the measurement cuvette.
- Luminometer measurements
Luminometer settings (96-well reader): Measurement duration per well = 1 sec (could be shorter, depending on the limits of your machine); cycle time might be set according to the number of total samples or used wells and the duration of one measurement.
Note: Some software/programs calculate the cycle time depending on the measurement duration and number of samples. Repetitions of measurement cycles/total time should be set to appropriately monitor signals for 20 min up to 60 min. It is recommended that in maximum only half of the plate is occupied with samples since the time difference between the measurement of the first and the last sample is too long. One round of measurement should be less than 60 sec. If you use a single cell luminometer you might just use the continuous mode or use a comparable program such as described above.
- Before the induction of a ROS-burst with MAMPs/elicitors, the background level has to be measured to ensure constant values of emitted light over time. Usually a measurement for about 5-10 min is sufficient to control for a stable non-oscillating baseline. If the baseline is not constant within this time the background measurement might be increased up to 60 min.
- Interrupt the background measurements and add your MAMP or any triggering substance (usually 1 µl) slightly shake plate horizontally on the table, and continue/re-start the measurement. The elicitors can be added by using an injector integrated in the machine, or by pipetting directly into the wells. Measurement cycles (30-60 sec, each) can be repeated to a total time of 20-60 min or longer if necessary.
- Before the induction of a ROS-burst with MAMPs/elicitors, the background level has to be measured to ensure constant values of emitted light over time. Usually a measurement for about 5-10 min is sufficient to control for a stable non-oscillating baseline. If the baseline is not constant within this time the background measurement might be increased up to 60 min.
Representative data
- Data handling and evaluation
The monitored ROS-burst is measured as emitted light due to the oxidation of luminol and is given in RLU (relative light units). Since RLU is not a clearly defined unit it can vary from machine to machine. Thus it is difficult to compare the results obtained with different machines. The measurement of the oxidative burst over time results in a kinetics curve as it is shown in the exemplary graph of Figure 1. For experiments it is important to use negative and positive controls, and leaf pieces of different plants should have the same size! Replicates (n ≥ 3) with equally sized leaf pieces are always necessary to perform statistics.
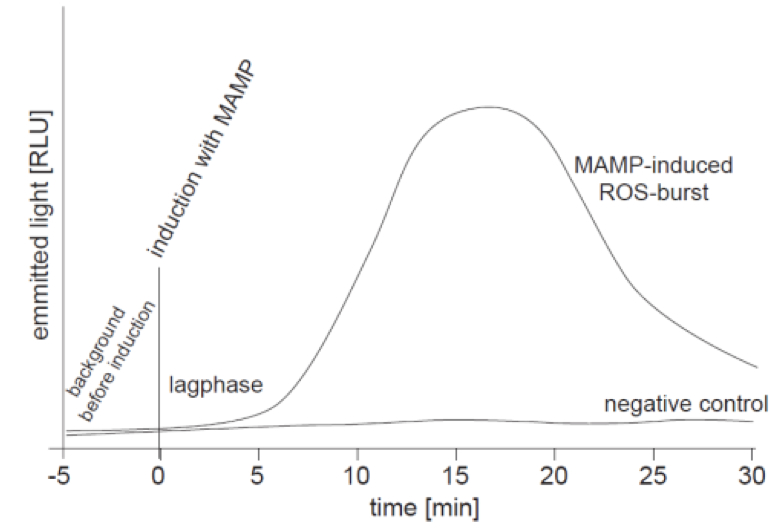
Figure 1. Schematic graph of a ROS-burst kinetics curve; ROS-burst was detected as emitted light in the luminol-based assay via a luminometer. Treatment of samples with a MAMP (e.g. flg22; 1 µl of a 10 µM stock solution; final concentration 100 nM) leads to a nice ROS-burst with a maximum peak at ~15 min. A lag-phase of 2-10 min is often observed. The negative control (e.g. water, BSA, etc.) does not cause a ROS-burst and the corresponding curve shows no peak.
To get an idea about the sensitivity of a system for a certain MAMP-trigger it is indispensable to measure the oxidative burst in a MAMP-dose-dependent manner. An appropriate amount of samples is treated with different doses of a MAMP, and the ROS-burst is recorded as described above. The obtained kinetics curves (examples in Figure 2 left panel) can be processed to a classical dose-response curve as shown in Figure 2 (right panel). In this way the EC50 can be determined that directly indicates the sensitivity of a biological system for a treatment with any trigger.
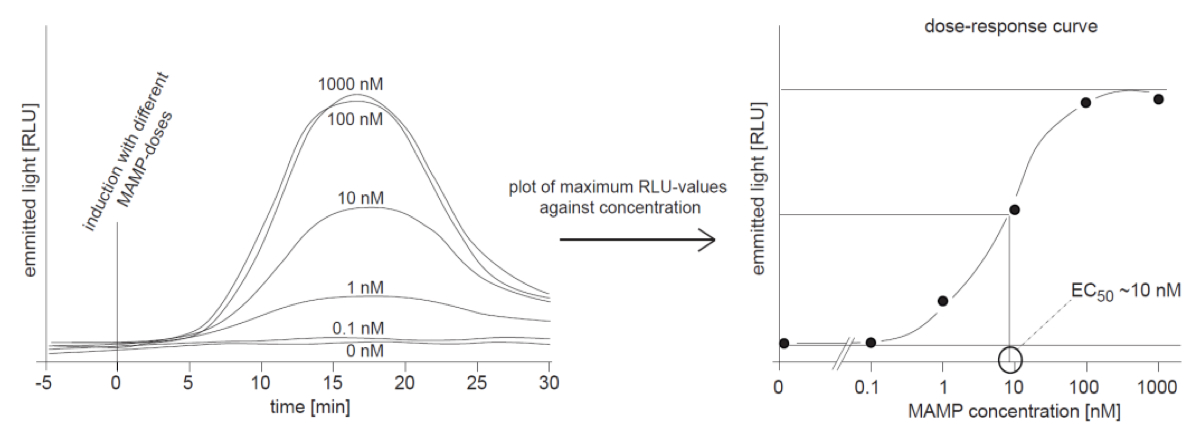
Figure 2. Dose-dependence of the ROS-burst response. Left panel: Samples were treated with different concentrations of a MAMP as indicated and ROS was recorded as emitted light over time. The highest concentrations (1,000 and 100 nM) give similar curves, indicating the system is saturated. The lower doses with concentrations of 10 and 1 nM result in curves with smaller amplitudes and a dose of 0.1 nM triggers no increase in ROS over the levels found in control treatment (water, 0 nM) Right panel: The maximum values from the left were plotted against the corresponding concentration and the graph shows a classical dose-response-curve. The EC50 of ~10 nM (half maximum effective concentration) was exemplary determined as indicated in the graph.
Recipes
- 10x luminol master mix
200 µM luminol L-012
10 µg/ml peroxidase, horseradish peroxidase
Acknowledgments
R.A.’s work was supported by Grant 348256/F20 from the Research Council of Norway; and Grant 216856 from the Research Council of Norway and the Deutscher Akademischer Austausch Dienst. M. A. was supported by the Deutsch Forschungsgemeinschaft (AL1426/1-1). The method was recently applied in Butenko et al. (2014).
References
- Albert, M., Jehle, A. K., Furst, U., Chinchilla, D., Boller, T. and Felix, G. (2013). A two-hybrid-receptor assay demonstrates heteromer formation as switch-on for plant immune receptors. Plant Physiol 163(4): 1504-1509.
- Albert, M., Jehle, A. K., Mueller, K., Eisele, C., Lipschis, M. and Felix, G. (2010). Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J Biol Chem 285(25): 19035-19042.
- Butenko, M. A., Wildhagen, M., Albert, M., Jehle, A., Kalbacher, H., Aalen, R. B. and Felix, G. (2014). Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26(5): 1838-1847.
- Felix, G., Duran, J. D., Volko, S. and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18(3): 265-276.
- Halter, T., Imkampe, J., Mazzotta, S., Wierzba, M., Postel, S., Bücherl, C., Kiefer, C., Stahl, M., Chinchilla, D. and Wang, X. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24(2): 134-143.
- Keppler, L. D., Baker, C. J. and Atkinson, M. M. (1989). Active oxygen production during a bacteria-induced hypersensitive reaction in tobacco suspension cells. Phytopathology 79(9): 974-978.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Albert, M., Butenko, M. A., Aalen, R. B., Felix, G. and Wildhagen, M. (2015). Chemiluminescence Detection of the Oxidative Burst in Plant Leaf Pieces. Bio-protocol 5(6): e1423. DOI: 10.21769/BioProtoc.1423.
Category
Plant Science > Plant immunity > Disease bioassay
Biochemistry > Other compound > Reactive oxygen species
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










