- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Gas Chromatography-Mass Spectrometry-Based Two Stage Assay for Measurement of in vitro myo-Inositol 3-phosphate Synthase (INO1) Activity
Published: Vol 5, Iss 5, Mar 5, 2015 DOI: 10.21769/BioProtoc.1418 Views: 10020
Reviewed by: Amit DeyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Radioactive-free Kinase Inhibitor Discovery Assay Against the Trypanosoma brucei Glycogen Synthase Kinase-3 short (TbGSK-3s)
Antonia Efstathiou and Despina Smirlis
Jan 20, 2020 4581 Views

Spectrophotometric Assessment of Heme Oxygenase-1 Activity in Leishmania-infected Macrophages
Moumita Basu [...] Anindita Ukil
Apr 5, 2020 5540 Views

In vitro Di-ubiquitin Formation Assay and E3 Cooperation Assay
Rebecca J. Burge [...] Jeremy C. Mottram
Nov 5, 2022 1878 Views
Abstract
This method describes an in vitro assay for measuring INO1 enzyme activity (the conversion of glucose 6-phosphate to myo-inositol 3-phosphate) in cell-free extracts. The method was first described for Plasmodium falciparum cells in MacRae et al. (2014) and consists of two parts: Part 1 describes the assay itself while part 2 describes analysis of the myo-inositol 3-phosphate product using gas chromatography-mass spectrometry (GC-MS).
Materials and Reagents
- Cells to be assayed [in the development of this protocol, we used Plasmodium falciparum (3D7 strain) cell cultures and human red blood cells (kindly supplied by the Australian Red Cross)]
- 13C-U-glucose (Cambridge Isotope Laboratories, catalog number: CLM1396 )
Note: 13C-U-glucose refers to Universally-labelled glucose - i.e. where all six carbons are 13C-atoms. - Hexokinase (>130 Units/mg) (Sigma-Aldrich, catalog number: H4502 )
- Adenosine triphosphate (ATP) (Sigma-Aldrich, catalog number: A6419 )
- Magnesium chloride (MgCl2, AnalaR) (VWR International, catalog number: 25108.260 )
- Tris-HCl (pH 7.5) (Sigma-Aldrich, catalog number: T5941 )
- Ammonium chloride (NH4Cl) (Ajax, catalog number: 31-500G )
- Nicotinamide adenine dinucleotide (NAD+) (Sigma-Aldrich, catalog number: N3014 )
- NaHEPES (pH 7.4) (Sigma-Aldrich, catalog number: H3375 )
- Ethylene glycol tetraacetic acid (Sigma-Aldrich, catalog number: E3889 )
- Dithiothreitol (Sigma-Aldrich, catalog number: D0362 )
- scyllo-Inositol (hereafter abbreviated to ‘sI’) (Sigma-Aldrich, catalog number: I8132 )
- Glucose 6-phosphate (Sigma-Aldrich, catalog number: G7879 )
- myo-Inositol 3-phosphate (Cayman Chemical Company, catalog number: CAY10007778 )
- Chloroform (HPLC grade) (Thermo Fisher Scientific, catalog number: 10615492 )
- Methanol (HPLC grade) (Thermo Fisher Scientific, catalog number: 10767665 )
- Assay buffer (see Recipes)
- Lysis buffer (see Recipes)
Additional materials required for GC-MS - Methoxyamine hydrochloride (Sigma-Aldrich, catalog number: 226904-25G )
- Pyridine (Sigma-Aldrich, catalog number: 270970 )
- BSTFA + 1% TMCS (Sigma-Aldrich, Supelco, catalog number: 33148 )
- Glucose (Sigma-Aldrich, catalog number: G8270 )
Equipment
- 1.5 ml tubes (with safe-lock lids) (Eppendorf)
- Bench-top centrifuge for 1.5 ml tubes
- 1 ml, 200 µl, and 20 µl pipettes and accompanying tips
- 37 °C water bath
- Boiling water bath
- Timer
- Distilled (e.g. MilliQ) water supply
- Light microscope, slides, and cover slips (for assessment of lysis)
- Bench-top vortex or water bath sonicator (may be required for efficient lysis)
Additional equipment required for GC-MS - Gas Chromatography-Mass Spectrometer (e.g. Agilent 7890B-5977A)
- DB-5MS + DG column, (30m x 0.25 mm, with 10 m inert gap) (Agilent, J&W)
- Ultra high purity helium
- 1.5 ml tubes (Eppendorf)
- 2 ml glass vials for mass spectrometry (e.g. Agilent, part number: 5182-0715 )
- 9 mm caps with septa for glass vials (e.g. Agilent, part number: 5185-5820 )
- 250 µl vial inserts (glass) (Agilent, part number: 5183-2085 )
- Rotary vacuum concentrator (e.g. Christ, model: RVC 2-33 CD Plus )
- Microdispensers and accompanying glass capillaries (50 µl adjustable, 200 µl) (Drummond Scientific Company)
Software
- ChemStation software (MSD ChemStation D.01.02.16) (Agilent)
Procedure
Note: An overview of the whole procedure can be seen in the accompanying figure.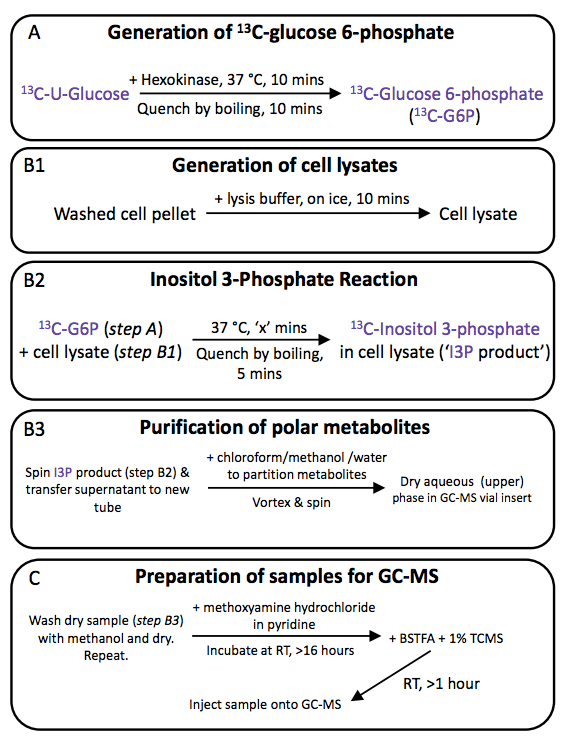
Figure 1. Inositol 3-phosphate synthase assay: overview
- Generation of 13C-glucose 6-phosphate
- 13C-glucose 6-phosphate is generated by incubation of [300 µM]final 13C-U-glucose with [66 µg ml-1]final hexokinase in assay buffer at 37 °C for 10 min, followed by boiling for 10 min in order to denature the hexose kinase. The tubes are then transferred to ice.
Note: We always produced the 13C-glucose 6-phosphate in bulk – enough for all reactions in the experiment. We reconstituted lyophilised hexokinase powder (>130 U/mg) in water at 10 mg/ml. This was added to the assay buffer at 6.8 per 1 ml assay buffer, to give a concentration of 68 μg/ml. A stock of 10 mM 13C-U-glucose was made (in water) and was added to the hexokinase/assay buffer at 30 μl per 1 ml, to give a final concentration of 300 μM 13C-U-glucose and 66 μg/ml hexokinase. Unused reaction products were stored at -80 °C for future use. - Efficiency of this conversion is usually ~100% and can be assessed by standard gas chromatography-mass spectrometry (GC-MS) protocols for sugar/sugar-phosphate analysis [see below, MacRae et al. (2014) and Saunders et al. (2011) for details].
- 13C-glucose 6-phosphate is generated by incubation of [300 µM]final 13C-U-glucose with [66 µg ml-1]final hexokinase in assay buffer at 37 °C for 10 min, followed by boiling for 10 min in order to denature the hexose kinase. The tubes are then transferred to ice.
- Generation of cell lysates and INO1 assay
- Lysates of your cells of choice are prepared by suspending washed cell pellets (with >10 pellet volumes of ice-cold PBS, two times) in ice-cold lysis buffer for 10 min.
Notes:- When measuring INO1 activity in Plasmodium falciparum-infected red blood cells, cells were suspended at a concentration of 109 cells per ml buffer (as determined by cell counting at the point of the second PBS wash). However, the volume of the suspension buffer may need to be varied depending on total cell number available and in vivo activity of INO1.
- The lysate should be checked (by light microscopy) to confirm successful lysis, some agitation (by either brief vortexing or sonication may be required).
- When measuring INO1 activity in Plasmodium falciparum-infected red blood cells, cells were suspended at a concentration of 109 cells per ml buffer (as determined by cell counting at the point of the second PBS wash). However, the volume of the suspension buffer may need to be varied depending on total cell number available and in vivo activity of INO1.
- Aliquots of 13C-glucose 6-phosphate substrate (50 µl) and cell lysate (50 µl) are combined, vortex mixed briefly (<1 sec), incubated in a water bath at 37 °C, and the reaction stopped at required time-points by boiling for 5 min. The tubes are then transferred to ice.
Note: In our experiments, we found that maximal labelling was achieved after ~3 h, although we also included later time points. A typical experiment would include 0, 0.5, 1, 5, 10, 30, 60, 120, 180, and 240 min, and an additional ‘overnight’ time point, if required. - Reaction solutions are then centrifuged at 16,100 x g and 4 °C for 5 min (to pellet cellular debris) and the supernatant (containing reaction products) transferred to a fresh 1.5 ml tube.
- Polar products are recovered by solvent extraction of 50 µl of the boiled assay mixture, following addition of chloroform (50 µl), methanol (150 µl), and MilliQ water (100 µl, containing 1 nmol sI as an internal standard), resulting in a final solvent ratio of chloroform/methanol/water (1:3:3 v/v/v). After vigorous vortex mixing (30 sec) and centrifugation at 16,100 x g and 4 °C for 5 min, this mixture results in two phases, a lower chloroform phase (~50 µl) and an upper methanol/water phase (~300 µl).
- Analysis of 13C-glucose 6-phosphate and the synthesised 13C-myo-inositol 3-phosphate can be easily quantified by GC-MS, by comparison to authentic standards, as described below and in MacRae et al. (2014) and Saunders et al. (2011).
- Lysates of your cells of choice are prepared by suspending washed cell pellets (with >10 pellet volumes of ice-cold PBS, two times) in ice-cold lysis buffer for 10 min.
- Sample preparation for gas chromatography-mass spectrometry (GC-MS)
- An aliquot of the upper polar phase (100 µl) is transferred to a GC-MS vial insert (placed in a 1.5 ml microfuge tube) and dried in a rotary vacuum concentrator (RVC). Once dry, an additional 100 µl of the polar phase is added to the insert and dried. This process is repeated until almost all of the upper phase has been transferred and dried.
Note: Be careful not to transfer any of the lipid-rich lower phase or the protein-rich interphase - it is advised to leave a few µl of the polar phase in the portioning tube rather than risk accidental transfer of the interphase and lower phase. Any minor differences in volume transferred are corrected later by normalisation to the internal standard (sI). - To ensure all water is displaced from the dried polar phase, 40 µl of methanol is added and dried in the RVC, followed by an additional 40 µl methanol and subsequent drying.
- The vial inserts are transferred to 2 ml glass vials with forceps, being careful not to touch the rim of the insert.
- Polar metabolites are methoximated by addition of a freshly prepared solution of 20 mg/ml methoxyamine in pyridine (20 µl, this should be prepared when required, i. e. while the second aliquot of methanol is being dried).
- The vials are capped, vortexed briefly (~5 sec) and incubated at RT for >16 h. The caps are removed and methoximated samples derivatised by addition of 20 µl of BSTFA + 1% TMCS, a reagent that results in trimethylsilyl (TMS) substitutions on the relevant metabolites, allowing them to be observable by GC-MS. The vials are re-capped, vortexed briefly and incubated at RT for >1 h before injection onto the GC-MS.
- An aliquot of the upper polar phase (100 µl) is transferred to a GC-MS vial insert (placed in a 1.5 ml microfuge tube) and dried in a rotary vacuum concentrator (RVC). Once dry, an additional 100 µl of the polar phase is added to the insert and dried. This process is repeated until almost all of the upper phase has been transferred and dried.
- Gas chromatography-mass spectrometry (GC-MS)
- Derivatised samples are analyzed using a DB-5MS + DG column in splitless mode (injection temperature 270 °C), or equivalent, using ultra high purity helium as the carrier gas.
- The initial oven temperature is 70 °C (2 min), followed by temperature gradients to 295 °C at 12.5 °C/min, and from 295 °C to 320 °C at 25 °C/min. The final temperature is held for 3 min.
- Data analysis can be performed using brand-specific specific or non-brand-specific software (e.g. AnalyzerPro, Amdis, mzMatch, mzMine, etc.). We used the accompanying ChemStation software.
- Metabolites are identified by comparison of retention times and ion fragmentation patterns with authentic standards. Quantification of metabolites is calculated using the formula: amount metabolite (nmol) = (area of appropriate metabolite peaks/area of sI peak) x (1 nmol sI/metaboliteMRRFsI); where metaboliteMRRFsI is the molar relative response factor determined from the mean value of: area of appropriate metabolite peak/area of sI peak (1:1 standards).
Notes:- The ‘appropriate peaks’ are the extracted ion chromatogram (EIC) peaks of appropriate unlabelled and labelled ions for each metabolite (see below).
- The level of labelling is estimated as the percent of the metabolite pool containing one or more 13C atoms, after background subtraction for naturally occurring isotopes (as calculated from an unlabelled standard), as described in Zamboni et al. (2009).
- The ‘appropriate peaks’ are the extracted ion chromatogram (EIC) peaks of appropriate unlabelled and labelled ions for each metabolite (see below).
- Ions used for quantification and label incorporation calculations:
scyllo-Inositol (sI, internal standard): M0 = m/z 318
myo-Inositol 3-phosphate: M0-M4 = m/z 318-322
Glucose: M0-M4 = m/z 319-323
Glucose 6-phosphate: M0-M2 = m/z 357-359
Note: M0 represents the unlabelled ion (where all carbon is 12C), Mn represents ions with n 13C-atoms.
- Derivatised samples are analyzed using a DB-5MS + DG column in splitless mode (injection temperature 270 °C), or equivalent, using ultra high purity helium as the carrier gas.
Recipes
- Assay buffer
1 mM ATP
2.5 mM MgCl2
100 mM Tris.HCl (pH 7.5)
14 mM NH4Cl
0.8 mM NAD+ - Lysis buffer
1 mM NaHEPES (pH 7.4)
2 mM ethylene glycol tetraacetic acid
2 mM dithiothreitol
Acknowledgments
This work was supported by a project grants from the National Health and Medical Research Council of Australia (NHMRC). M.J.M is an NHMRC Principal Research Fellow and J.I.M. was supported by a Royal Society Travelling Fellowship.
References
- MacRae, J. I., Lopaticki, S., Maier, A. G., Rupasinghe, T., Nahid, A., Cowman, A. F. and McConville, M. J. (2014). Plasmodium falciparum is dependent on de novo myo‐inositol biosynthesis for assembly of GPI glycolipids and infectivity. Mol Microbiol 91(4): 762-776.
- Saunders, E. C., Ng, W. W., Chambers, J. M., Ng, M., Naderer, T., Kromer, J. O., Likic, V. A. and McConville, M. J. (2011). Isotopomer profiling of Leishmania mexicana promastigotes reveals important roles for succinate fermentation and aspartate uptake in tricarboxylic acid cycle (TCA) anaplerosis, glutamate synthesis, and growth. J Biol Chem 286(31): 27706-27717.
- Zamboni, N., Fendt, S. M., Ruhl, M. and Sauer, U. (2009). (13)C-based metabolic flux analysis. Nat Protoc 4(6): 878-892.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
MacRae, J. I. and McConville, M. J. (2015). A Gas Chromatography-Mass Spectrometry-Based Two Stage Assay for Measurement of in vitro myo-Inositol 3-phosphate Synthase (INO1) Activity. Bio-protocol 5(5): e1418. DOI: 10.21769/BioProtoc.1418.
Category
Microbiology > Microbial biochemistry > Carbohydrate
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









