- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Splenic Dendritic Cells Using Fluorescence-activated Cell Sorting
Published: Vol 5, Iss 5, Mar 5, 2015 DOI: 10.21769/BioProtoc.1415 Views: 17901
Reviewed by: Kathrin SutterAndrea IntroiniAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2243 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views
Abstract
The spleen is a vastly vasculated organ and consists of a complex organized network of innate and adaptive immune cells. This permits the specialized functions of the spleen such as antibacterial and antifungal immunity and iron metabolism among others (Mebius and Kraal, 2005). Different dendritic cell (DC) subsets reside in the spleen and can be defined by the expression of unique surface markers. These DC subsets are recognized to perform non-redundant functions in the immune system (Merad et al., 2013). In our recent study, we found that Inositol Requiring Enzyme (IRE)-1 is specifically activated in splenic CD8a+ DCs. Furthermore, loss of X-box binding protein (XBP)-1 – the transcription factor regulated by IRE-1 – resulted in defective cross-presentation of dead cell associated antigens by splenic CD8a+ DCs (Osorio et al., 2014). This protocol allows the isolation of specific DC subsets for experimental use ex-vivo.
Materials and Reagents
- Mice (Jackson Laboratories, C57Bl/6 )
- 2,2,2-tribromoethanol (Sigma-Aldrich, catalog number: T48402 )
- 2-methyl-2-butanol (Sigma-Aldrich, catalog number: 240486 )
- PBS (Gibco, catalog number: 10010-015 )
- 0.5 M EDTA (Lonza, catalog number: 51234 )
- 2-mercaptoethanol (Sigma-Aldrich, catalog number M3148 )
- BSA (Amresco, catalog number: 0332-500g )
- HBSS (Life Technologies, catalog number: 24020-091 )
- RPMI 1640 GlutaMax (Life Technologies, catalog number: 61870-010 )
- Gentamycin (Gibco, catalog number: 15710-049 )
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F7524 )
- Liberase TM (Roche Diagnostics, catalog number: 0 5401127001 ) (dilute 50 mg in 50 ml of RPMI, aliquot and stored at -20 °C)
- DNase I (Roche, catalog number: 0 4536282001 ) (dilute content in 1 ml PBS, aliquot and stored at -20 °C)
- FACS antibodies
- CD3 - FITC (eBioscience, clone 145-2C11, dilution 1/300)
- CD19 - FITC (eBioscience, clone 1D3, dilution 1/500)
- CD11c - PE-eFluor610 (eBioscience, N418, dilution 1/300)
- MHCII - APC-Cy7 (Biolegend, clone M5/114.15.2, dilution 1/1,000)
- CD8a - PE-Cy5 (BD-Pharmingen, clone 53-6.7, dilution 1/600)
- CD11b - PE-Cy7 (BD-Pharmingen, clone M1/70, dilution 1/800)
- CD64 - AF647 (BD-Pharmingen, clone X54-5/7.1, dilution 1/100)
- CD16/CD32 (Produced in-house, clone 2.4G2, dilution 1/200)
Note: The use of 2.4G2 minimizes background fluorescence by inhibiting non-specific binding of antibodies to FcG – receptors present on immune cells.
- CD3 - FITC (eBioscience, clone 145-2C11, dilution 1/300)
- Fixable viability dye eFluor 506 (eBioscience, catalog number: 65-0866-18 )
- Reconstitute vial with 400 μl PBS
- Aliquot and store vials at -80 °C
- Use at a dilution of 1/200
- Reconstitute vial with 400 μl PBS
- Anti-FITC microbeads (Miltenyi, catalog number 130-048-701 )
- LD columns (Miltenyi, catalog number: 130-042-901 )
- Ultra comp eBeads (eBioscience, catalog number: 01-2222-42 )
- Trypan blue (Sigma-Aldrich, catalog number: T8154 )
- Avertin (see Recipes)
- 2x digestion medium (see Recipes)
- MACS buffer (see Recipes)
- R10 medium (see Recipes)
- Osmotic lysis buffer (see Recipes)
Equipment
- 100 μm Nylon cell strainer (Falcon, catalog number: 352360 )
- 15 ml tubes (Falcon, catalog number: 430791 )
- 35 mm Petri Dishes
- 5ml syringe (Norm-Ject, catalog number: 4020-000V0 )
- 25 Gauge needle (Terumo, catalog number: NN-2516R )
- Pasteur pipette
- Forceps
- Surgical scissor/scalpel
- 37 °C warm water bath
- Shaker
- Centrifuge
- Aspiration device
- MACS MultiStand (Miltenyi, catalog number: 130-042-303 )
- Midi MACS (Miltenyi, catalog number: 130-042-302 )
- Polystyrene 5 ml tubes with 100 μm cell strainer cap (BD Falcon, catalog number: 352235 )
- Polypropylene 5ml tubes (BD Falcon, catalog number: 352063 )
- Microscope
- Bürker-Türk counting plate
- Multicolour FACS sorter (BD Aria II or equivalent cell sorter equipped with 405 nm, 488 nm and 633 nm lasers and appropriate filterset)
Procedure
- Euthanize the mice by injecting 0.5 ml of avertin intraperitoneally with a 25 Gauge needle. The mice become unresponsive after 1-2 min.
Note: Verify the institution approved ethical regulations on animal welfare if other euthanization methods are in place.
- Check the absence of cerebrospinal reflexes by pinching the paw of the posterior limbs with a forceps.
- After spraying the animal with 70% ethanol, open the right flank and remove the spleen. Remove any fatty tissue that is connected with the spleen and place the organ in 15 ml tube with 5 ml of ice-cold HBSS.
- Bring the spleen to a 35 mm Petri dish and mince the organ into pieces by using the scissors. To obtain a good dissociation of the tissue, proceed until no pieces are visible with the bare eyes.
- Add 1 ml of ice-cold RPMI, bring the cell suspension to a 15 ml Falcon tube and put on ice. Complete all other organs before adding the digestion medium.
- Add 1 ml of 37 °C preheated 2x digestion medium to the cell suspension and bring all organs to the 37 °C warm water bath. Make sure the shaker is put to its maximum setting.
Note: Shaking the sample will allow more efficient digestion of the tissue by decreasing its precipitation.
- Incubate the cell suspension for 15 min and dissociate the cell suspension by pipetting the solution up and down through a Pasteur pipette.
- Repeat step 7 once more.
- Add 10 ml of ice-cold MACS buffer and spin down cell suspension at 400 x g 7 min 4 °C.
- Aspirate the supernatant and resuspend the cell pellet in 2 ml of ice-cold osmotic lysis buffer. Incubate for 4’ at room temperature and subsequently add 10 ml of ice-cold MACS buffer. Spin down cell suspension at 400 x g 7 min 4 °C.
- Resuspend cells in 10 ml of ice-cold MACS buffer and filter through a 100 μm cell strainer. One can reuse the 15 ml tube to collect the filtered cells. Take a 20 μl counting sample and spin down rest of cell suspension at 400 x g 7 min 4 °C.
- Add 180 μl of trypan blue to the 20 μl of counting sample and mix well. Count cells. After digestion, 1 spleen will normally contain up to 60-100 x106 cells.
- Remove supernatant from cell pellet and resuspend in antibody mix I at a concentration of 50 x 106 cells per 1 ml of mix (see table 1). Stain 20 min on ice in the dark. Add 10 ml of ice-cold MACS buffer and spin down 7 min 400 x g 4 °C.
Table 1. Antibody mix I in MACS buffer
Label
Marker
Clone
Dilution
FITC
CD3
145-2C11
1/300
FITC
CD19
1D3
1/500
-
CD16/CD32
2.4G2
1/200
- Remove supernatant from cell pellet and add 90 μl of ice-cold MACS buffer per 10 x 106 cells.
- Add 5-10 μl/10 x 106 cells of anti-FITC microbeads. Resuspend and incubate 15 min on ice. Add 10 ml of ice-cold MACS buffer and spin down 7 min 400 x g 4 °C. Discard supernatant.
- Prepare LD columns according to manufacturer’s protocol.
- After preparation, place a 15 ml tube on ice under the LD column.
- Resuspend cell pellet in 1 ml of ice-cold MACS buffer and filter through a 100 μm cell strainer on the LD column.
Note: This minimizes the probability of cell clots obstructing the LD column. After the 1 ml of cell suspension has run through, wash 2 times with 2 ml of ice-cold MACS buffer. Make sure that cell suspension runs through each time. Discard the column and spin down the effluent (7 min 400 x g 4 °C). Aspirate the supernatant.
- Stain cell pellet in 500 μl of antibody mix II. Stain 20 min on ice in the dark. Add 10 ml of ice-cold MACS buffer and spin down 7 min 400 x g 4 °C. Aspirate supernatant and resuspend in 1 ml of ice-cold MACS buffer. Bring the cell suspension to a 5 ml polystyrene tube with a cell strainer cap to remove possible clots.
Table 2. Antibody mix II in PBS
Note: Use of PBS is necessary for optimal staining of dead cells with the Fixable viability dye eFluor 506.Label
Marker
Clone
Dilution
FITC
CD3
145-2C11
1/300
FITC
CD19
1D3
1/500
PE-eFluor610
CD11c
N418
1/300
PE-Cy5
CD8a
53-6.7
1/600
PE-Cy7
CD11b
M1/70
1/800
AF-647
CD64
X54-5/7.1
1/100
APC-Cy7
MHCII
M5/114.15.2
1/1,000
eFluor
506
Viability
1/200
-
CD16/CD32
2.4G2
1/200
- Single stains are prepared by diluting 1 drop of Ultra comp eBeads in 2 ml of PBS. Make sure to shake the beads thoroughly before use as precipitation occurs. Bring 200 μl of this bead suspension to seven 5 ml reaction tubes (6 fluorochromes + 1 unstained sample). Antibodies conjugated to bright fluorochomes (PE-eFluor610, Pe-Cy5, AF-647) are diluted 1/2,000, other antibodies are diluted 1/400. Stain 15 min on ice in the dark. Add 1 ml of PBS and spin down 7 min 400 x g 4 °C. Remove supernatant and resuspend beads in 200 μl of PBS. Proceed to the cell sorter.
Note: Viability dyes do not bind to the Ultra comp eBeads. Since we exclude dead cells (defined by a positive staining for eFluor 506) there is no need to compensate this fluorochrome. If compensation is preferred, cells can be stained with a 1 in 200 dilution of the eFluor 506 viability dye in PBS. Stain 20 min on ice in the dark. Wash cells with 1ml of PBS and spin down 7 min 400 x g 4 °C. Resuspend the cells in 200 μl of PBS.
- An example of a gating setup is shown below.
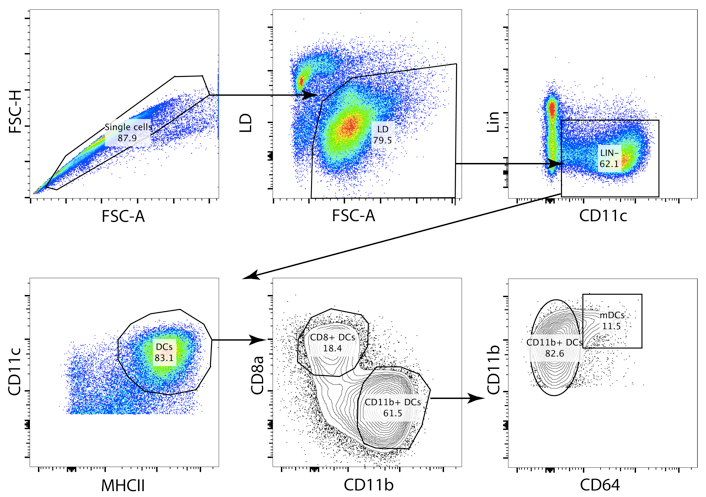
Figure 1. Gating strategy of splenic DCs. A strict gating strategy in the FSC-H/FSC-A plot allows the exclusion of doublets. The CD11b+ DCs (Lin- CD11c+ MHCII+ CD11b+ cells) contain a fraction of monocyte derived cells (specific infections with pathogens or inflammation can drastically increase this cell population, contaminating the CD11b+ DC gate). The conventional CD11b+ DCs can be identified by exclusion of the CD64+ cells.
Notes
- One should take in mind to work as fast as possible. Once removed from the live animal splenic DC subsets viability is quickly compromised. Furthermore, working with cold reagents improves overall survival.
- A good dissociation of the tissue by use of scissor and Pasteur pipettes and adequate incubation of the splenocytes in the digestion medium is crucial for optimal cell retrieval. When adjusting the antibody mix, keep in mind that the enzymatic digestion can potentially cleave off surface proteins expressed on the cell types of interest.
- In optimal conditions, negative selection with the use of MACS beads removes 90-95% of CD3e+ and CD19+ cells. This reduces the number of cells to approximate 5-10 x 106 cells and a total sorting time to approximate 20 min.
- One can sort 80,000-120,000 CD8a+ DCs and 400,000-600,000 CD11b+ DCs from 1 spleen (in homeostatic conditions). The CD11b+ DCs can be further subdivided by the use of surface markers ESAM or CD4.
- In the case that the cells are to be cultured, the scientist should keep in mind to work sterile from the moment the spleen is obtained (step 4). Prior to use, the buffers can be filtered to remove potential pathogens using a 0.2 μm filter.
- Cells can be brought into culture in R10 medium with the addition of 250 ng/ml of recombinant murine Flt3L. Without any further stimulation; survival of these dendritic cell subsets is less than 60% after 24 h of culture.
Recipes
- Avertin (for 40 ml)
Dissolve 1 g of 2,2,2-tribromoethanol in 1 ml of 2-methyl-2-butanol
Resuspend in 39 ml of PBS and shake over night at room temperature
Avoid direct exposure to light and store at 4 °C
- 2x digestion medium
Reconstitute RPMI 1640 with:
1/500 DNAse I (10 IU/ml)
1/25 liberase TM (0.42 Wünsch Unit/ml)
- MACS buffer
1x PBS
2% FCS (or 2% BSA)
5 mM EDTA
Note: This minimizes adherence of cells to plastics by chelating Ca2+.
Filter sterilize over a 0.2 μm filter and store at 4 °C
- R10 medium
Reconstitute RPMI1640-Glutamax with:
10% FBS
50 μg/ml gentamycin
50 μM beta-mercaptoethanol
- Osmotic lysis buffer (500 ml total)
450 ml MilliQ water (or other source of ultra-pure ddH2O)
4.145 g ammonium chloride
100 μl 500 mM EDTA
Adjust pH to 7.1-7.4
Add MilliQ water up to 500 ml
Filter sterilize over a 0.2 μm filter and stored at 4 °C
Acknowledgments
This work was supported by the European Research Council, the European Union Seventh Framework Programme, the Fonds Wetenschappelijk Onderzoek Vlaanderen program (FWO) and the Agentschap voor Innovatie door Wetenschap en Technologie (IWT).
References
- Merad, M., Sathe, P., Helft, J., Miller, J. and Mortha, A. (2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 31: 563-604.
- Mebius, R. E. and Kraal, G. (2005). Structure and function of the spleen. Nat Rev Immunol 5(8): 606-616.
- Osorio, F., Tavernier, S. J., Hoffmann, E., Saeys, Y., Martens, L., Vetters, J., Delrue, I., De Rycke, R., Parthoens, E., Pouliot, P., Iwawaki, T., Janssens, S. and Lambrecht, B. N. (2014). The unfolded-protein-response sensor IRE-1alpha regulates the function of CD8alpha+ dendritic cells. Nat Immunol 15(3): 248-257.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tavernier, S. J., Osorio, F., Janssens, S. and Lambrecht, B. N. (2015). Isolation of Splenic Dendritic Cells Using Fluorescence-activated Cell Sorting. Bio-protocol 5(5): e1415. DOI: 10.21769/BioProtoc.1415.
Category
Immunology > Immune cell isolation > Antigen-presenting cell
Immunology > Immune cell staining > Flow cytometry
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










