- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Degradation Assays in Arabidopsis Protoplasts
Published: Vol 5, Iss 4, Feb 20, 2015 DOI: 10.21769/BioProtoc.1397 Views: 16507
Reviewed by: Zhaohui LiuCindy AstAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification of S-locus F-box Protein Sequences in Diploid Potato, Solanum okadae, via Degenerate PCR
Amar Hundare [...] Timothy P. Robbins
Jun 5, 2025 2005 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2025 Views

Isolation and Transfection of Protoplasts From Maize Mesophyll Cells
Lauren A. Higa [...] Zhi-Yan Du
Feb 5, 2026 100 Views
Abstract
Plant transformation and exogenous protein expression is essential for molecular biology and biotechnology. Current approaches of stable plant transformation might be problematic and very time-consuming. Because of this, transient expression in protoplasts has become valuable alternative, being less cost and time-effective at the same time. Excellent for eukaryotic proteins, representing a natural cell habitat, protoplast isolation is widely used in protein interaction visualization techniques, like BiFC (Bimolecular fluorescence complementation) and FRET (Förster resonance energy transfer). In this protocol we present a another use of Arabidopsis protoplast in protein degradation assay, proving its high versatility as a tool in proteomics.
Materials and Reagents
- 3-week old Arabidopsis plants
- Mannitol (BDH Prolabo, catalog number: 25311 )
- CaCl2 (POCH, catalog number: M00015143 )
- KCl (USB, catalog number: 20598 )
- 2-(N-morpholino)ethanesulfonic acid (MES) (LabEmpire, catalog number: MES503 )
- NaCl (POCH, catalog number: BA4121116 )
- Polyethylene glycol (PEG) 4000 (Sigma-Aldrich, catalog number: 81240 )
- Cellulase (SERVA Electrophoresis GmbH, catalog number: 16419 )
- Macerozyme (SERVA Electrophoresis GmbH, catalog number: 28302 )
- GenEluteTM HP Plasmid Midiprep Kit (Sigma-Aldrich, catalog number: PLD35 )
- Benzyloxycarbonyl-L-leucyl-L-leucyl-L-leucinal, Z-Leu-Leu-Leu-al (MG132) (Sigma-Aldrich, catalog number: M7449 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418 )
- HEPES buffer (Sigma-Aldrich, catalog number: H3375 )
- MgCl2 (USB, catalog number: 18641 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
- Protease inhibitors (Roche Diagnostics, catalog number: 11873580001 )
- Enzyme solution (see Recipes)
- W5 (see Recipes)
- W1 (see Recipes)
- MMg (see Recipes)
- 40% PEG (see Recipes)
- Protein isolation buffer (see Recipes)
Equipment
- Light microscope (Nikon Corporation)
- Hemocythometer (Sigma-Aldrich)
- Tape (Scotch® MagicTM)
- Stainless steel forceps (Sigma-Aldrich)
- Scissors
- 10 ml pipette
- 50 ml tubes
- 90 mm Petri dishes
- Amicon Ultra-15 centrifugal filter unit (Millipore)
- NanoDrop spectrophotometer
- Tabletop centrifuge
- Horizontal shaker
Procedure
- Protoplast isolation
- Using tweezers, 5-6 mature leaves of Arabidopsis, petioles removed, were harvested.
- Adaxial surface of leaves was put on a piece of tape and leaves were flattened (Figure 1A-B).
- Second piece of tape was applied with limited pressure, trapping leaves between them (Figure 1C). To much pressure will result in damaging leaves, too little pressure will lead to poor epidermis removal effectiveness.
- Sandwich was placed with adaxial surface on top and pieces of tape were split by pulling away tape on top side, started at the tip of leaf (Figure 1D-E) (see Note 2).
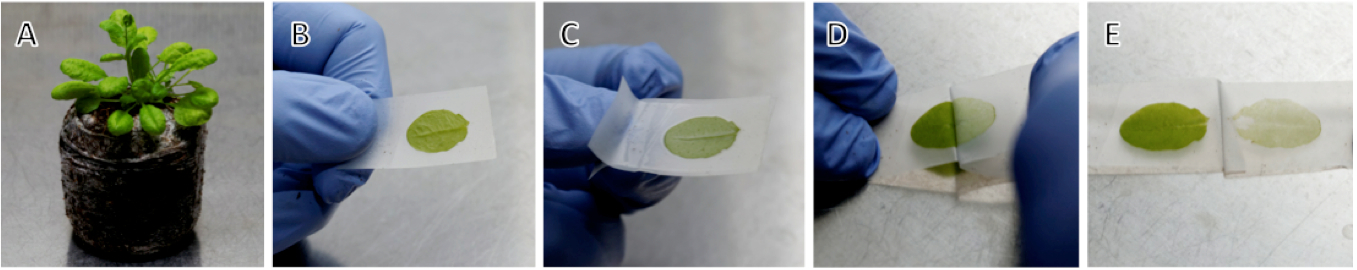
Figure 1. Preparation of Arabidopsis leaf for protoplast isolation. A. 3-weeks old Aradodopsis thaliana; B-E. Step by step procedure of leaf epidermis removal with tape. - Tape was cut around leaves and put at room temperature enzyme solution in Petri dish, exposed mesophyll down. Tape should not be immersed in solution, but float on top of it. Mesophyll cells were digested for 60 min at 30 °C with gentle shaking (55 rpm) (see Note 3).
- After digestion pieces of tape were removed and cells were left for another 5 min in same conditions (see Note 4).
- Using 10 ml pipette protoplasts were transported to 50 ml tubes and put on ice.
- Protoplast were centrifuged for 3 min (150 x g, 4 °C) and washed twice with W5 buffer. Be careful not to resuspend protoplast to abruptly. After second wash step protoplast were resuspended in 1 ml of MMg solution. Protoplasts were calculated with help of hemocythometer and diluted to optimal concentration of 2 x 104 cells in 100 μl with MMg solution (see Notes 5-7).
- 5-10 μg plasmid DNA coding a tag-protein fusion was alequoted to 2 ml eppendorf tube (see Note 8).
- 100 μl of isolated protoplasts in MMg medium were transferred to the tube using a pipette tip with the tip of the tip cut off.
- Using a pipette tip with the tip of the tip cut off 110 μl of PEG solution was added. Solution was gently mixed and left for 15 min at RT in horizontal position (see Note 9).
- After incubation, 450 μl of W1 buffer was added to the tubes to dilute PEG solution, mixed and centrifuged for 3 min, 300 x g.
- Supernatant was removed and harvested protoplasts were resuspended in 300 μl of W1 solution.
- Transfected protoplasts were incubated overnight at 22 °C in horizontal position in the dark for protein expression. It is important not to disturb protoplasts at this stage.
- From this point protoplasts can be used in various protein analysis techniques (kinase assay, in vivo protein degradation assay, protein subcellular localization, BiFC and FRET analyses) (Ludwikow et al., 2014). Here we show in vivo degradation assay for plant proteins.
- Using tweezers, 5-6 mature leaves of Arabidopsis, petioles removed, were harvested.
- In vivo degradation assay in protoplasts
- After overnight incubation (as indicated in step A14) in the dark protoplasts were treated with 50 μM MG132 (proteasome inhibitor) or mock treated with 0.1% DMSO for 6 h.
- After brief centrifugation (300 x g, RT) supernatant was removed and cells were resuspended and disrupted in 100 μl of protein isolation buffer.
- Protein concentration was determined using a NanoDrop spectrophotometer.
- Prepared samples were separated by SDS-PAGE and further analyzed by Western blotting.
- After overnight incubation (as indicated in step A14) in the dark protoplasts were treated with 50 μM MG132 (proteasome inhibitor) or mock treated with 0.1% DMSO for 6 h.
Representative data
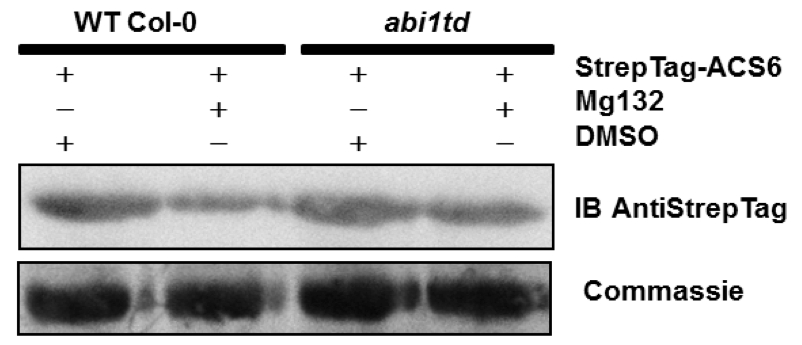
Figure 2. MG132 treatment increases ACC synthase6 protein accumulation in the abi1td protoplasts
Protoplasts isolated from WT Col-0 and the ABI1 knockout line (abi1td) were transformed with 5 µg of DNA plasmid coding for StrepTag-ACS6. Transformed protoplasts were treated with 50 µM MG132 or an equivalent volume of DMSO (mock control) for 6 h prior harvesting. A Western blot with anti-StrepTag antibodies confirms the presence of the StrepTag-ACS6 protein. The Western blot shown is representative of at least three independent experiments. Coomassie staining confirms equal protein loading (Ludwikow et al., 2014).
Notes
- This protocol is applicable to Brassica napus protoplast isolation and transformation.
- To avoid mesophyll cells damage (visible as dark green spots) don't use too much pressure when applying the tape.
- It is not recommended to digest leaves for more than 60 min. Protoplasts yield at this point will not increase, but they lose viability.
- If one hour digestion did not freed all mesophyll cells from tape fragments, one can gentle dip tape a few times in enzyme solution, to increase protoplast yield.
- Use swinging bucket rotor for centrifugation.
- Keep low acceleration and deceleration values during centrifugation.
- Resuspend the protoplasts by gently rocking the tube.
- Use hemocytometer to achieve accurate and reproducible results.
- A large volume of plasmid DNA decreases transformation efficiency, therefore keep the volume around 10 μl. Low transformation efficiency is usually a result of low quality plasmid DNA.
Recipes
- Enzyme solution (10 ml)
1.2% cellulose
0.4% macerozyme
0.4 M mannitol
20 mM KCl
20 mM MES
dH2O
Filter sterilize
Incubate at 55 °C for 10 min
Prepare fresh, do not store - W5 (50 ml)
154 mM NaCl
125 mM CaCl2
5 mM KCl
2 mM MES
dH2O
Filter sterilize, autoclave
Stored at 4 °C - W1 (10 ml)
0.5 M mannitol
20 mM KCl
4 mM MES
dH2O
Filter sterilize, autoclave
Stored at 4 °C, but no longer than 2 weeks - MMg (10 ml)
0.4 M mannitol
15 mM MgCl2
4 mM MES
dH2O
Filter sterilize, autoclave
Stored at 4 °C, but no longer than 2 weeks - 40% PEG (10 ml)
4 g PEG 4000
200 mM mannitol
100 mM CaCl2
Filter sterilize, autoclave
Stored at 4 °C, but no longer than 3 weeks - Protein isolation buffer
20 mM HEPES (pH 7.5)
10 mM MgCl2
1 mM DTT
1 mM PMSF
Protease inhibitor
dH2O
Stored at -20 °C
Acknowledgments
This work was supported by COST Action FA0605 project 682/N-COST/2010/0, the National Science Centre grants (5615/B/P01/2010/39, DEC-2012/05/B/NZ3/00352, DEC-2011/03/N/NZ3/01796) and POLAPGEN grant no. WND-POIG.01.03.01-00-101/08. This protocol was adapted from Wu et al. (2009).
References
- Ludwików, A., Ciesla, A., Kasprowicz-Maluśki, A., Mitula, F., Tajdel, M., Galganski, L., Ziolkowski, P. A., Kubiak, P., Malecka, A., Piechalak, A., Szabat, M., Gorska, A., Dabrowski, M., Ibragimow, I. and Sadowski, J. (2014). Arabidopsis protein phosphatase 2C ABI1 interacts with type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Mol Plant 7(6): 960-976.
- Wu, F. H., Shen, S. C., Lee, L. Y., Lee, S. H., Chan, M. T. and Lin, C. S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mituła, F., Kasprowicz-Maluśki, A., Michalak, M., Marczak, M., Kuczyński, K. and Ludwików, A. (2015). Protein Degradation Assays in Arabidopsis Protoplasts. Bio-protocol 5(4): e1397. DOI: 10.21769/BioProtoc.1397.
Category
Plant Science > Plant cell biology > Cell isolation
Plant Science > Plant molecular biology > Protein
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











