- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Phosphatase Protection Assay: 14-3-3 Binding Protects the Phosphate group of RSG from λ Protein Phosphatase
Published: Vol 5, Iss 3, Feb 5, 2015 DOI: 10.21769/BioProtoc.1395 Views: 10705
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Split-luciferase Complementation Imaging Assay to Study Protein-protein Interactions in Nicotiana benthamiana
Liping Wang [...] Rosa Lozano-Durán
Dec 5, 2021 12064 Views

In vitro Auto- and Substrate-Ubiquitination Assays
Hye Lin Park [...] Gyeong Mee Yoon
Apr 5, 2022 3524 Views

Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
Anna Karlsson [...] Elton P. Hudson
May 5, 2025 2011 Views
Abstract
14-3-3 proteins regulate diverse cellular processes in eukaryotes by binding to phospho-serine or threonine of target proteins. One of the physiological functions of 14-3-3 is to bind and protect phosphate groups of the target proteins against phosphatases. REPRESSION OF SHOOT GROWTH (RSG) is a tobacco (Nicotiana tabacum) transcription factor that is involved in the feedback regulation of biosynthetic genes of plant hormone gibberellin. 14-3-3 binds to phospho-Ser-114 in RSG. Ca2+-dependent protein kinase NtCDPK1 was identified as a kinase that phosphorylates Ser-114 of RSG. Our recent study revealed that NtCDPK1 forms a heterotrimer with RSG and 14-3-3 and that 14-3-3 was transferred from NtCDPK1 to phosphorylated RSG (Ito et al., 2014). In the course of the study, we found that 14-3-3 protects the phosphate group of RSG from λ protein phosphatase in vitro. Here, we describe a protocol for in vitro phosphatase protection assay. To detect the phosphorylation state of proteins, we used Phos-tag SDS-PAGE and autoradiography. This protocol can be adapted for the examinations whether the phosphoprotein-binding proteins protect the phosphate group of target proteins from phosphatases although protein kinases may be required for the phosphorylation of target proteins.
Keywords: PhosphataseMaterials and Reagents
- 20 mM ATP
- [γ-32P] ATP (5,000 Ci/mmol, 10 mCi/ml) (Institute of Isotopes, catalog number: SBP-401 )
- λ protein phosphatase (New England Biolabs, catalog number: P0753S )
- Amylose resin (New England Biolabs, catalog number: E8021 )
- COSMOGEL His-Accept (Nacalai Tesque, catalog number: 08368-12 )
- Glutathione Sepharose 4B (GE Healthcare, catalog number: 17-0756-01 )
- Phos-tag acrylamide (Wako Pure Chemical Industries, catalog number: 304-93521 )
- Wide-view prestained protein size marker III (Wako Pure Chemical Industries, catalog number: 230-02461 )
- His.tag monoclonal antibody (Merck Millipore, catalog number: 70796-4CN )
- Phenylmethanesulfonyl fluoride (PMSF)
- Dialysis buffer (see Recipes)
- 10x phosphorylation buffer (see Recipes)
- Binding buffer (see Recipes)
- Dephosphorylation buffer (see Recipes)
- 2x SDS protein sample buffer (see Recipes)
Equipment
- 1.5 ml microcentrifuge tubes
- Refrigerated microcentrifuge
- Microtube rotator
- GELoader tips 0.5-20 µl (Eppendorf, catalog number: 0030 001.222 )
- Heat block
- SDS-PAGE apparatus
- Gel transfer apparatus
- Power supply
- Shaker
- Phosphorimager
- Chemiluminescence imaging system
Procedure
- Preparation of recombinant proteins
- MBP-RSG, His-14-3-3 and GST-NtCDPK1 were expressed in Escherichia coli containing pMALc2-RSG, pET16b-14-3-3 or pGEX 4T-1-NtCDPK1, respectively, as described in the references (Ishida et al., 2008; Ito et al., 2010; Ito et al., 2014).
- MBP-RSG, His-14-3-3 and GST-NtCDPK1 were purified by using Amylose resin, COSMOGEL His-Accept or Glutathione Sepharose 4B according to the manual (pMALTM Protein Fusion & Purification System, Ni-NTA His Bind Resins, or GST gene fusion system handbook, respectively).
- The purified proteins were dialyzed separately overnight against a 500-1,000-fold volume of ice-cold dialysis buffer.
- Glycerol, corresponding to 40% by volume of the dialyzed protein solutions, was added to the dialyzed protein solutions to make approximately 50% (v/v) glycerol (final concentration), and the protein solutions were stored at -20 °C.
Note: Dialyzed protein solution contains approximately 30% (v/v) glycerol.
- MBP-RSG, His-14-3-3 and GST-NtCDPK1 were expressed in Escherichia coli containing pMALc2-RSG, pET16b-14-3-3 or pGEX 4T-1-NtCDPK1, respectively, as described in the references (Ishida et al., 2008; Ito et al., 2010; Ito et al., 2014).
- Phosphorylation of RSG by NtCDPK1
- Prepare reaction mixtures in each of 1.5 ml microcentrifuge tube using the following:
5 µl 10x phosphorylation buffer
5 µl 1% (v/v) β-mercaptoethanol (dilute immediately before use)
2.5 µl 20 mM ATP with or without 1 µl [γ-32P] ATP (5,000 µCi/mmol, 10 mCi/ml)
2 µg MBP-RSG
100 ng GST-NtCDPK1
H2O up to 50 µl
- Mix and incubate the reaction mixture for 1 h at 30 °C.
- Prepare reaction mixtures in each of 1.5 ml microcentrifuge tube using the following:
- 14-3-3 binding to phosphorylated RSG
- Gently invert the container of amylose resin to resuspend the slurry.
- Transfer 30 µl of resuspended slurry to each of 1.5 ml microcentrifuge tube.
- Add 1 ml of ice-cold binding buffer to each microcentrifuge tube to equilibrate the resin for use.
- Spin down the resin at 1,000 x g for a few seconds at 4 °C.
- Carefully remove and discard the supernatant.
- Repeat steps C3-5 two more times for a total of three washes.
- Add 250 µl of ice-cold binding buffer, all (50 µl) of the reaction mixture phosphorylated RSG and 3 µg His-14-3-3 to the resin.
- Gently mix the mixture for 30 min at 4 °C on a microtube rotator to bind MBP-RSG to the amylose resin and His-14-3-3.
- Spin down the resin at 1,000 x g for a few seconds at 4 °C.
- Carefully remove and discard the supernatant.
- Wash the resin with 1 ml of ice-cold binding buffer to remove ATP and unbound proteins.
- Gently mix by inverting the tube several times.
- Spin down the resin at 1,000 x g for a few seconds at 4 °C.
- Carefully remove and discard the supernatant.
- Repeat steps C11-14 three more times for a total of four washes.
- Completely remove the supernatant by using a micropipettor equipped with GELoader Tips.
- Gently invert the container of amylose resin to resuspend the slurry.
- Dephosphorylation of RSG by λ protein phosphatase
- Add 200 µl of ice-cold dephosphorylation buffer and 1 µl (400 U) of λ protein phosphatase to the resin.
- Gently mix the mixture for 30 min at 4 °C on a microtube rotator.
- Wash unbound proteins from the resin with 1 ml of ice-cold binding buffer.
- Gently mix by inverting the tube several times.
- Spin down the resin at 1,000 x g for a few seconds at 4 °C.
- Carefully remove and discard the supernatant.
- Repeat steps D3-6 three more times for a total of four washes.
- Completely remove the supernatant by using a micropipettor equipped with GELoader tips.
- Add 200 µl of ice-cold dephosphorylation buffer and 1 µl (400 U) of λ protein phosphatase to the resin.
- Detection of phosphorylation state of RSG
- Add 60 µl of 2x SDS protein sample buffer to the resin.
- Heat the tubes for 5 min in a 95 °C heat block to denature the proteins.
- Load 25 µl of the supernatant and 3 µl of protein size marker and perform Phos-tag SDS-PAGE (Kinoshita and Kinoshita-Kikuta, 2011) for the detection of MBP-RSG phosphorylated with cold ATP or normal SDS-PAGE for the detection of MBP-RSG phosphorylated with [γ-32P] ATP.
- Perform coomassie brilliant blue (CBB) staining of Phos-tag SDS-PAGE gel to examine the phosphorylation state of MBP-RSG phosphorylated with cold ATP.
- To detect the phosphorylation state of MBP-RSG phosphorylated with [γ-32P] ATP, dry the SDS-PAGE gel and place an imaging plate over the dried gel. Expose overnight to visualize the radioactive signals. Scan the imaging plate by using an imaging system, Typhoon FLA 9500.
- Perform immunoblot analysis with anti-14-3-3 (Igarashi et al., 2001) antibody or anti-His-tag monoclonal antibody for the detection of His-14-3-3 bound to MBP-RSG by using an imaging system, ImageQuant LAS 4000.
- Add 60 µl of 2x SDS protein sample buffer to the resin.
Representative data
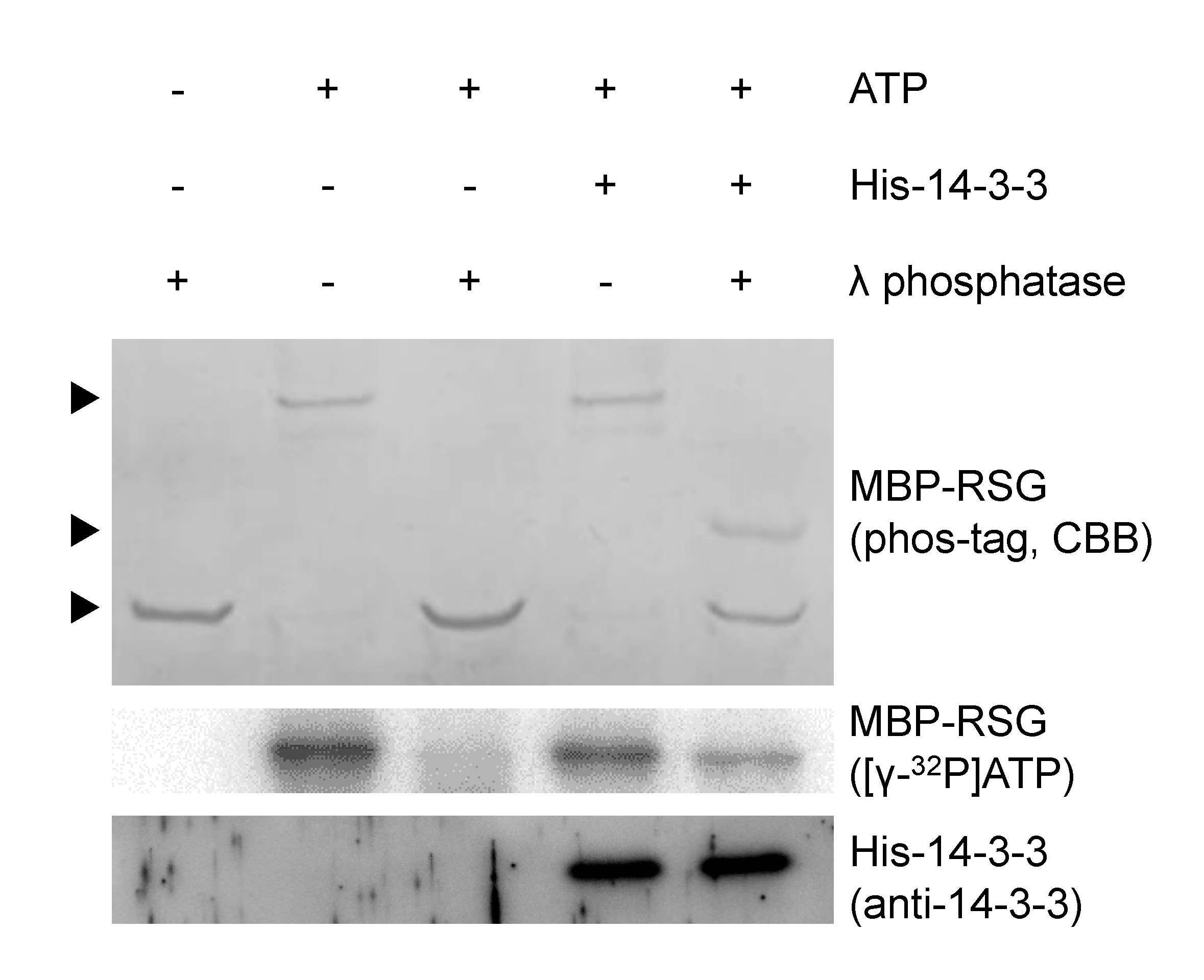
Figure 1. The phosphorylation state of Ser-114 in RSG is protected from λ protein phosphatase by 14-3-3. MBP-RSG proteins phosphorylated with cold ATP were resolved on Phos-tag SDS-PAGE and visualized by coomassie brilliant blue (CBB) staining. MBP-RSG proteins phosphorylated with [γ-32P] ATP were resolved on SDS-PAGE and detected by using imaging system. Several amino acids of RSG are phosphorylated by NtCDPK1. When His-14-3-3 does not bind to MBP-RSG, phosphate groups of RSG are completely removed by λ protein phosphatase. On the other hand, when MBP-RSG that bound to His-14-3-3 is treated with λ protein phosphatase, the phosphorylation state of Ser-114 in RSG is protected from λ protein phosphatase. Top, middle and bottom arrowheads represent multiply phosphorylated, Ser-114 phosphorylated and dephosphorylated MBP-RSG, respectively. For more information, please see Ito et al. (2014).
Notes
- We used λ protein phosphatase but not bacterial alkaline phosphatase for this assay because 14-3-3 tends to dissociate from RSG under alkaline conditions. If alkaline conditions do not lead to the dissociation of target protein complexes, bacterial alkaline phosphatase can be used for this assay.
- The condition for stable protein-protein interactions vary among different proteins. The optimum buffer condition for target protein-protein interactions could be found by changing pH level, NaCl (KCl) concentration and detergent type and concentration.
- In case that phosphorylation or dephosphorylation is excessive or insufficient, changing the reaction times or the amounts of kinase and phosphatase could gain satisfactory results.
Recipes
- Dialysis buffer
25 mM Tris-HCl (pH 7.5 at 4 °C)
150 mM NaCl
0.1% (v/v) Triton X-100
30% (v/v) glycerol
0.05% (v/v) β-mercaptoethanol
0.1 mM PMSF
Note: β-mercaptoethanol and PMSF are added immediately before use.
- 10x phosphorylation buffer
200 mM Tris-HCl (pH 7.5 at room temperature)
100 mM NaCl
1% (v/v) Triton X-100
5 mM CaCl2
- Binding buffer
25 mM MOPS-NaOH (pH 7.0 at 4 °C)
25 mM NaCl
0.1% (v/v) Triton X-100
0.05% (v/v) β-mercaptoethanol
0.1 mM PMSF
Note: β-mercaptoethanol and PMSF are added immediately before use.
- Dephosphorylation buffer
25 mM MOPS-NaOH (pH 7.0 at 4 °C)
25 mM NaCl
0.1% (v/v) Triton X-100
1 mM MnCl2
0.05% (v/v) β-mercaptoethanol
0.1 mM PMSF
Note: β-mercaptoethanol and PMSF are added immediately before use.
- 2x SDS protein sample buffer
250 mM Tris-HCl (pH 6.8 at room temperature)
4.0% (w/v) SDS
20% (v/v) glycerol
0.02% (w/v) Bromophenol blue
10% (v/v) β-mercaptoethanol
Note: β-mercaptoethanol is added immediately before use.
Acknowledgments
This work was supported by the Japan Society of the Promotion of Science (grant no. 23657038 to Y. T.) and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant no. 24118004 to Y. T.).
References
- Igarashi, D., Ishida, S., Fukazawa, J. and Takahashi, Y. (2001). 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13(11): 2483-2497.
- Ishida, S., Yuasa, T., Nakata, M. and Takahashi, Y. (2008). A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20(12): 3273-3288.
- Ito, T., Nakata, M., Fukazawa, J., Ishida, S. and Takahashi, Y. (2010). Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca(2+)-dependent protein kinase is important for substrate recognition. Plant Cell 22(5): 1592-1604.
- Ito, T., Nakata, M., Fukazawa, J., Ishida, S. and Takahashi, Y. (2014). Scaffold function of Ca2+-dependent protein kinase: Tobacco Ca2+-DEPENDENT PROTEIN KINASE1 transfers 14-3-3 to the substrate REPRESSION OF SHOOT GROWTH after phosphorylation. Plant Physiol 165(4): 1737-1750.
- Kinoshita, E. and Kinoshita-Kikuta, E. (2011). Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11(2): 319-323.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ito, T. and Takahashi, Y. (2015). Phosphatase Protection Assay: 14-3-3 Binding Protects the Phosphate group of RSG from λ Protein Phosphatase. Bio-protocol 5(3): e1395. DOI: 10.21769/BioProtoc.1395.
- Ito, T., Nakata, M., Fukazawa, J., Ishida, S. and Takahashi, Y. (2014). Scaffold function of Ca2+-dependent protein kinase: Tobacco Ca2+-DEPENDENT PROTEIN KINASE1 transfers 14-3-3 to the substrate REPRESSION OF SHOOT GROWTH after phosphorylation. Plant Physiol 165(4): 1737-1750.
Category
Plant Science > Plant biochemistry > Protein > Interaction
Biochemistry > Protein > Interaction > Protein-protein interaction
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










