- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection and Quantification of Heme and Chlorophyll Precursors Using a High Performance Liquid Chromatography (HPLC) System Equipped with Two Fluorescence Detectors
Published: Vol 5, Iss 3, Feb 5, 2015 DOI: 10.21769/BioProtoc.1390 Views: 13038
Reviewed by: Tie LiuSeda Ekici

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Extraction and Analysis of Carotenoids from Escherichia coli in Color Complementation Assays
Andreas Blatt and Martin Lohr
Mar 20, 2017 12245 Views

Evaluation of the Condition of Respiration and Photosynthesis by Measuring Chlorophyll Fluorescence in Cyanobacteria
Takako Ogawa and Kintake Sonoike
May 5, 2018 8255 Views

Analysis of Metals in Whole Cells, Thylakoids and Photosynthetic Protein Complexes in Synechocystis sp. PCC6803
Chiara Gandini [...] Sidsel Birkelund Schmidt
Jun 20, 2018 6340 Views
Abstract
Intermediates of tetrapyrrole biosynthetic pathway are low-abundant compounds, and their quantification is usually difficult, time consuming and requires large amounts of input material. Here, we describe a method allowing fast and accurate quantification of almost all intermediates of the heme and chlorophyll biosynthesis, including mono-vinyl and di-vinyl forms of (proto) chlorophyllide, using just a few millilitres of the cyanobacterial culture. Extracted precursors are separated by High Performance Liquid Chromatography system (HPLC) and detected by two ultra-sensitive fluorescence detectors set to different wavelengths.
Keywords: Chlorophyll biosynthesisMaterials and Reagents
- Methanol (HPLC grade)
- Pyridine reagent Plus >99% (Sigma-Aldrich, catalog number: P57506 )
- H2O (HPLC grade) (Merck KGaA)
- Acetonitrile (HPLC grade)
- HPLC solvent A (see Recipes)
- HPLC solvent B (see Recipes)
Equipment
- 0.5 ml polypropylene tubes
- HPLC 2 ml glass vials and 200 μl glass inserts
- HPLC system with an autosampler (Agilent, model: 1200 ) (Figure 1)
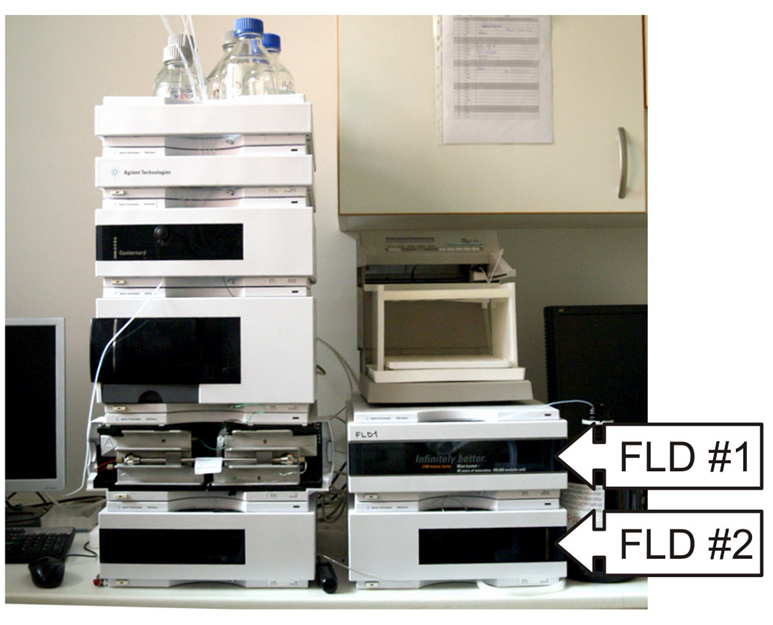
Figure 1. The Agilent 1200 HPLC system equipped with an autosampler and two fluorescence detectors marked as FLD #1 and FLD #2 - Two HPLC fluorescence detectors (Agilent, model: 1200 ) (see Note 1)
The original Agilent 1200 detectors offer adequate sensitivity for emission wavelengths ranging 300-600 nm, however for longer wavelengths the sensitivity quickly falls almost to zero. This issue also applies to the newer 1260 version of the detector. As sensitivity to longer wavelengths is essential for detection of several heme/chlorophyll precursors, the original Agilent photomultiplier was replaced with the R928 photomultiplier tube (Hamamatsu, catalog number: R928) in both detectors. This modification significantly improved sensitivity within the entire wavelength range, and dramatically improved the sensitivity to wavelengths >600 nm. The replacement for the photomultiplier was provided by Agilent’s technical service. - Tabletop centrifuge (Eppendorf, MiniSpin plus)
- Reverse phase column (ReproSil 100 C8-AB, 5-µm particle size, 4 x 250 mm) (Dr. Maisch, catalog number: r15.8b.s2504 )
Procedure
- The protocol requires 2 ml of a culture of cyanobacterial cells at the exponential phase of growth. The protocol works very well for the cyanobacterium Synechocystis PCC 6803 grown at 30 °C in the liquid medium BG11 (Rippka et al., 1979) with optical density at 750 nm ~ 0.4 (see Note 2). The BG11 medium is used frequently for various cyanobacterial strains, it contains a balanced mix of macro- and microelements required for photoautotrophic grow and its preparation is described in detail in Rippka et al. (1979).
- All further steps should be carried out under dim or green light to prevent degradation of photo labile tetrapyrroles. Alternatively, the tubes can be covered by aluminum foil.
- Harvest cells by centrifuging at room temperature and maximal speed (≥ 12 000 x g) for 5 min. Remove the supernatant and resuspend cells in 200 µl of ultrapure H2O. Transfer the cell solution into a 500 µl tube, spin cells down again and carefully remove all water by pipette. At this point, cells can be frozen in liquid nitrogen and stored in -70 °C for up to a month.
- Resuspend the pellet from the end of step 3 in 25 µl of ultrapure water, then add 75 µl of methanol, mix thoroughly and incubate in dark and at room temperature for 15 min to extract tetrapyrroles. Pellet cell debris by centrifuging for 5 min at maximal speed.
- Transfer the supernatant (~ 90 µl) to a new 500 µl tube.
- Resuspend the pellet from the end of step 3 in 20 µl of ultrapure water, add 80 µl of methanol, mix thoroughly and incubate again in dark and at room temperature for 15 min. Pellet cell debris by centrifuging for 5 min at maximal speed.
- Pool both supernatants (~ 180 µl) in the 500 µl tube, mix thoroughly and centrifuge again for 5 min at maximal speed to remove any traces of cell debris. Transfer the supernatant to the 200 µl insert and inject it immediately into the HPLC system.
- Separation is performed on a reverse phase column with 35% methanol and 15% acetonitrile in 0.25 M pyridine (solvent A) and 50% methanol in acetonitrile as solvent B. Tetrapyrroles are eluted with a gradient of solvent B (35 to 50% in 5 min) followed by 50 to 64% of solvent B in 30 min at a flow rate of 0.8 ml/min at 40 °C (Table 1). The program continues by column washing (36-45 min) and equilibration (36-45 min).
Table 1. Setting of HPLC pumps for a single runTime (min) % of the solvent B 0 35 5 50 35 64 36 100 45 100 46 35 51 35 - Each run is finished using 100% of solvent B for 10 min to wash out more hydrophobic and abundant pigments like chlorophylls of carotenoids. The column is finally equilibrated for 5 min by 35 % of the solvent B (Table 1 for the complete pump program).
- Eluted pigments are detected by their specific fluorescence emission maxima. Settings of fluorescence detectors, the elution times and the excitation/emission maxima of individual precursors are summarized in Table 1 and also indicated graphically in Figure 2.
Table 2. Settings of fluorescent detectors and the list of detectable heme/chlorophyll precursors, including their excitation and emission maxima and approximate elution times
a-divinylDetector Detector Setting Ex/Em (nm), Time Precursor, Ex/Em maxima (nm) Elution time FLD #1 400/620, 0-10 min Coproporphyrin IX 400/420 6 min 440/660, 11-22 min DVa-chlorophylide 440/670 17 min MVb-chlorophylide 440/670 18 min DV -protochlorophylide 440/640 20 min MV-protochlorophylide 440/640 21 min 400/630, 13-30 min Protoporphyrin IX 400/630 26 min FLD #2 416/595, 0-30 min Mg-protoporphyrin IX 416/595 18 min Mg-protoporphyrin IX MEc 416/595 24 min
b-monovinyl
c-monomethyl-ester
Representative data

Figure 2. Representative chromatograms of separated precursors extracted from the cyanobacterium Synechocystis PCC 6803. A. Chromatogram recorded by the first fluorescent detector (FLD). B. Chromatogram recorded simultaneously by the second FLD. Settings of excitation and emission wavelengths for FLD #1 and #2 are shown above the each chromatogram. The number in round brackets indicates the FLD sensitivity (gain; the maximum = 16) set for the given wavelengths (see Note 3). Copro III = Coproporphyrin IX; DV-PChlide = divinyl-protochlorophyllide; MV-PChlide = monovinyl-protochlorophyllide; DV-Chlide = divinyl-chlorophyllide; MV-Chlide = monovinyl-chlorophyllide; Proto IX - Protoporphyrin IX; Mg-Proto = Mg-protoporphyrin IX; Mg-Proto ME = Mg-protoporphyrin IX monomethylester.
Notes
- The protocol should work using any HPLC system with binary or quaternary pump system and with at least one fluorescence detector sensitive enough for the longer wavelengths (up to 680 nm). To detect the whole spectrum of precursors using a system with one fluorescence detector, each sample needs to be measured twice with different setting of excitation/emission wavelengths.
- Apart from the cyanobacterium Synechocystis PCC 6803 the described protocol has already been successfully tested for the green alga Chlamydomonas reinhartii, the moss Physcomitrella patens, the plant Arabidopsis thaliana, and for Escherichia coli. Coproporphyrin IX and Protoporphyrin IX should be detectable virtually in any organism.
- Sensitivity of the Agilent fluorescence detector can be set from 1-16 (max). This particular setting (numbers) is specific for the Agilent detectors but a similar increasing/decreasing of the sensitivity should be possible to achieve with virtually any HPLC fluorescence detector.
Recipes
- HPLC solvent A
35% methanol and 15% acetonitrile in 0.25 M pyridine - HPLC solvent B
50% methanol in acetonitrile
Acknowledgments
This research project was supported by projects Algatech (CZ.1.05/2.1.00/03.0110) and Algain (EE2.3.30.0059) of the Czech Ministry of Education, Youth and Sports.
References
- Knoppova, J., Sobotka, R., Tichy, M., Yu, J., Konik, P., Halada, P., Nixon, P. J. and Komenda, J. (2014). Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26(3): 1200-1212.
- Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M. and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111(1): 1-61.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pilný, J., Kopečná, J., Noda, J. and Sobotka, R. (2015). Detection and Quantification of Heme and Chlorophyll Precursors Using a High Performance Liquid Chromatography (HPLC) System Equipped with Two Fluorescence Detectors. Bio-protocol 5(3): e1390. DOI: 10.21769/BioProtoc.1390.
Category
Plant Science > Plant physiology > Photosynthesis
Plant Science > Plant metabolism > Other compound
Biochemistry > Other compound > Chlorophyll
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









