- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Intracellular Cytokine Staining on PBMCs Using CyTOFTM Mass Cytometry
Published: Vol 5, Iss 1, Jan 5, 2015 DOI: 10.21769/BioProtoc.1370 Views: 24131
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for Screening Host-Targeting Antivirals (HTAs) Using Human PBMCs and pDCs
Zhao Xuan Low [...] Pouya Hassandarvish
Mar 5, 2025 3068 Views

Assessing Human Treg Suppression at Single-Cell Resolution Using Mass Cytometry
Jonas Nørskov Søndergaard [...] James B. Wing
Aug 20, 2025 2923 Views

Dual Phospho-CyTOF Workflows for Comparative JAK/STAT Signaling Analysis in Human Cryopreserved PBMCs and Whole Blood
Ilyssa E. Ramos [...] James M. Cherry
Nov 20, 2025 2351 Views
Abstract
In this protocol, we use a CyTOFTM mass cytometry to collect single-cell data on a large number of cytokines/chemokines as well as cell-surface proteins that characterize T cells and other immune cells. The current selected mass window in AW 103-203 includes the lanthanides used for most antibody labeling, along with iridium and rhodium for DNA intercalators. The output data are in the format as .txt and .fcs files, which is compatible with many analysis programs. This protocol could be adapted to include tetramers into the staining panel, but we have not optimized for that purpose.
The principal steps of intracellular cytokine staining are as follows: First, cells are activated for a few hours using either a specific peptide or a non-specific activation cocktail. An inhibitor of protein transport (e.g. Brefeldin A) is added to retain the cytokines within the cell. Next, EDTA is added to remove adherent cells from the activation vessel. After washing, antibodies to cell surface markers are added to the cells. The cells are then fixed in paraformaldehyde and permeabilized. We use a gentle detergent, saponin, as the permealization buffer because it is less destructive to surface and intracellular epitopes compared to harsh detergents or methanol. After permeabilization, the metal-conjugated anti-cytokine antibodies are added into the cell suspension. The stained cells are then sequentially introduced into the mass cytometry for signal intensity analysis.
Materials and Reagents
- PBMC (fresh or thawed frozen)
- RPMI-1640 (Hyclone, catalog number: SH30027.01 )
- FBS (Atlanta Biologicals, catalog number: S11150 )
- Pen-strep-Glutamin 100X (Hyclone, catalog number: SV30082.01 )
- Benzonase (2.5 x 105 U/ml) (Pierce, catalog number: 88701 )
- Brefeldin A (Sigma-Aldrich, catalog number: B7651 )
- Monensin (Sigma-Aldrich, catalog number: M5273 )
- 0.5 M EDTA (Hoefer, catalog number: GR-123-100 )
- Sodium azide (10% w/v solution) (Teknova, catalog number: S0209 )
- 16% para-formaldehyde (PFA) (Alfa Aesar, catalog number: 43368 ))
- 10x PBS (ROCKLAND, catalog number: MB-008 )
- BSA (Sigma-Aldrich, catalog number: A7284 )
- Maleimide-DOTA (Macrocyclics, catalog number: B-272 )
- Lanthanum (III) chloride heptahydrate (Sigma-Aldrich, catalog number: 203521 )
- Indium (III) chloride (Sigma-Aldrich, catalog number: 203440 )
- MilliQ water
Note: Beakers or bottles used here are not washed with soap due to barium content of most commercial soaps.
- Phenotyping antibodies (filtered with 0.1 µm spin filters) (Millipore, catalog number: UFC30VV00 )
- Ir-intercalator stock solution (Fluidigm, catalog number: 201192 )
Note: Rh103-intercalator can be used.
- 10x saponin-based permeabilization buffer (eBioscience, catalog number: 00 8333-56 )
- Phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich, catalog number: P8139 )
- Ionomycine (Sigma-Aldrich, catalog number: I0634 )
- Phytohemagglutinin (PHA) (Sigma-Aldrich, catalog number: 61764 )
- SEB (Sigma-Aldrich, catalog number: S0812 )
- Anti-CD3/CD28 (various vendors)
- Peptide mixes (JPT)
- Complete RPMI (see Recipes)
- CyPBS (see Recipes)
- CyFACS buffer (see Recipes)
- Live-dead stain (see Recipes)
Equipment
- 96- well round-bottom plates
- 37 °C water bath
- Biosafety cabinet
- Centrifuge
- CO2 incubator at 37 °C
- Calibrated pipettes
Procedure
- Thaw PBMC
- Warm complete RPMI media to 37 °C in water bath. Each sample will require 22 ml of media with benzonase. Calculate the amount needed to thaw all samples, and prepare a separate aliquot of warm media with 1:10,000 benzonase (final concentration 25 U/ml). Benzonase is added into the media to prevent dead cell aggregation. Thaw no more than 3 samples at a time. Run one control PBMC with each batch of samples.
- Remove samples from liquid nitrogen and transport to lab on dry ice.
- Place 10 ml of warmed benzonase media into a 15 ml tube, making a separate tube for each sample.
- Thaw frozen vials in 37 °C water bath.
- When cells are nearly completely thawed, carry to hood.
- Add 1 ml of warm benzonase media from appropriately labeled centrifuge tube slowly to the cells, then transfer the cells to the centrifuge tube. Rinse vial with more media from centrifuge tube to retrieve all cells.
- Continue with the rest of the samples as quickly as possible.
- Centrifuge cells at 1,550 rpm (RCF = 473) for 8 min at room temperature.
- Remove supernatant from the cells and resuspend the pellet by tapping the tube.
- Gently resuspend the pellet in 1 ml warmed benzonase media. Filter cells through a 70 micron cell strainer if needed. Add 9 ml more warmed benzonase media to the tube.
- Centrifuge cells at 1,550 rpm (RCF = 473) for 8 min at room temperature. Remove supernatant from the cells and resuspend the pellet by tapping the tube.
- Resuspend cells in 1 ml warm media.
- Count cells with Vicell (or hemocytometer if necessary). To count, take 20 μl cells and dilute with 480 μl PBS in Vicell counting chamber. Load onto Vicell as PBMC with a 1:25 dilution factor.
- Adjust the cell concentration to 5-10 x 106 cells/ml with warm media (no more benzonase at this point).
- Using a multichannel pipette, add 200 µl cells (for at least 1 x 106 cells) into each well of a 96-well deep well plate. Split each sample equally into two or more wells keeping one as an unstimulated control and the others for different types of stimulation.
- Rest overnight (6-18 h) at 37 °C in CO2 incubator.
- Warm complete RPMI media to 37 °C in water bath. Each sample will require 22 ml of media with benzonase. Calculate the amount needed to thaw all samples, and prepare a separate aliquot of warm media with 1:10,000 benzonase (final concentration 25 U/ml). Benzonase is added into the media to prevent dead cell aggregation. Thaw no more than 3 samples at a time. Run one control PBMC with each batch of samples.
- Stimulate cells
- After overnight rest at 37 °C, add the activation reagents and secretion inhibitor (brefeldin A/monensin) to the well for stimulation. Add only the secretion inhibitor to the unstimulated control well. If doing CD107a staining, add CD107a antibody during the stimulation.
Table 1. Protein secretion inhibitors
Reagent
Stock concentration
Intermediate dilution
Final concentration
Brefeldin A
5 mg/ml in DMSO
(stored in aliquots at -20 °C)
1:10 in PBS
10 μg/ml (1:50) or 5 μg/ml (1:100) with monensin
Monensin
5 mg/ml in ethanol
(stored at -20 °C)
1:10 in PBS
10 μg/ml (1:50) or 5 μg/ml (1:100) with brefeldinA
Table 2. Activators
Reagent
Stock concentration
Intermediate dilution
Final concentration
Phorbol 12-myristate 13-acetate (PMA)
1 mg/ml in DMSO
(stored in aliquots at -20 °C)
1:1,000 in PBS
10 ng/ml
Ionomycine
1 mg/ml in DMSO
(stored in aliquots at -20 °C)
1: 10 in PBS
1 µg/ml
Phytohemagglutinin (PHA)
1 mg/ml in DMSO
(stored at 4 °C)
1:10 in PBS
1 µg/ml
SEB
50 μg/ml in PBS
None
1 μg/ml (1:50)
Anti-CD3/CD28
Follow manufacturer instruction
-
-
Peptide mixes
0.5-1 mg/ml/pep in DMSO
(stored in aliquots at -20 °C)
1:10 in PBS
1 μg/ml/peptide
(1:50 - 1:100)
Notes:
- It is important to avoid solvent toxicity. Final DMSO+ethanol concentration from all sources (peptides, brefeldin A, monensin) should not exceed 0.5%.
- For most cytokines: Use brefeldin A at 10 μg/ml final concentration (see stock preparation table). For CD107 and CD154: Use monensin at 10 μg/ml final concentration (see stock preparation table). For assays combining cytokines and CD107 or CD154: Use brefeldin A and monensin at 5 μg/ml final concentration each.
- Metal-labeled CD107 and CD154 can be added into the culture during the stimulation at a concentration of 2 μg/ml. This allows for staining of target molecules that are re-internalized by cells during the activation process.
- Addition of costimulatory antibodies is optional. Add 1 μg/ml final concentration of CD28 and/or CD49d (labeled antibody can be used if analysis of the marker is desired).
- It is important to avoid solvent toxicity. Final DMSO+ethanol concentration from all sources (peptides, brefeldin A, monensin) should not exceed 0.5%.
- Incubate the cells for 4 h (PMA + ionomycin stimulation, PHA + ionomycin stimulation) or 6-8 h (SEB, anti-CD3/CD28 stimulation, peptide stimulation) at 37 °C, in a CO2 incubator.
Note: For most cytokines 6-12 h incubation at 37 °C is sufficient; For IL-10 optimal incubatation time is 12-24 h, but it is possible to detect in 6 h.
- At the end of stimulation, add EDTA to a final concentration of 2 mM and incubate for 15 min at room temperature.
- After overnight rest at 37 °C, add the activation reagents and secretion inhibitor (brefeldin A/monensin) to the well for stimulation. Add only the secretion inhibitor to the unstimulated control well. If doing CD107a staining, add CD107a antibody during the stimulation.
- Staining
- Wash 2x with 250 µl CyFACS buffer per well 1,550 rpm (RCF = 473), 10 min at room temperature. The same volume and centrifuge conditions are used in additional wash steps before fixation with PFA (step C6).
- Make surface Ab cocktail in CyFACS buffer (filter with 0.1 µm spin filter) 100 µl per reaction. Incubate on ice for 45 min. Use vendor’s recommended concentration (or optimal titer as determined for self-made conjugates) for each antibody.
- Wash 2x in CyFACS buffer.
- Resuspend cells in 100 µl of 1:3,000 diluted 5 mg/ml 115-DOTA maleimide (titrated if new stock) in CyPBS, Incubate 30 min on ice.
- Wash 3x in CyFACS buffer.
- Resuspend in 100 µl of 2% PFA in CyPBS, Incubate 4 °C overnight.
- Wash 2x in 1x eBioscience perm buffer (1x in milliQ water), 2,000 rpm (RCF = 787), 10 min at 4 °C. The same volume and centrifuge conditions are used in the following wash steps.
- Make intracellular staining cocktail in 1x perm buffer and filter with 0.1 µm spin filter, 100 µl per reaction. Incubate on ice for 45 min.
- Wash 3x in CyFACS buffer.
- Resuspend in 100 µl 1: 2,000 Ir-Interchelator diluted in 2% PFA (in CyPBS).
- Incubate 20 min at room temp.
- Wash 2x in CyFACS buffer.
- Wash 3x in MilliQ water.
- Resuspend in MilliQ water (1 to 1.5 ml) for running on CyTOF.
- Wash 2x with 250 µl CyFACS buffer per well 1,550 rpm (RCF = 473), 10 min at room temperature. The same volume and centrifuge conditions are used in additional wash steps before fixation with PFA (step C6).
Representative data
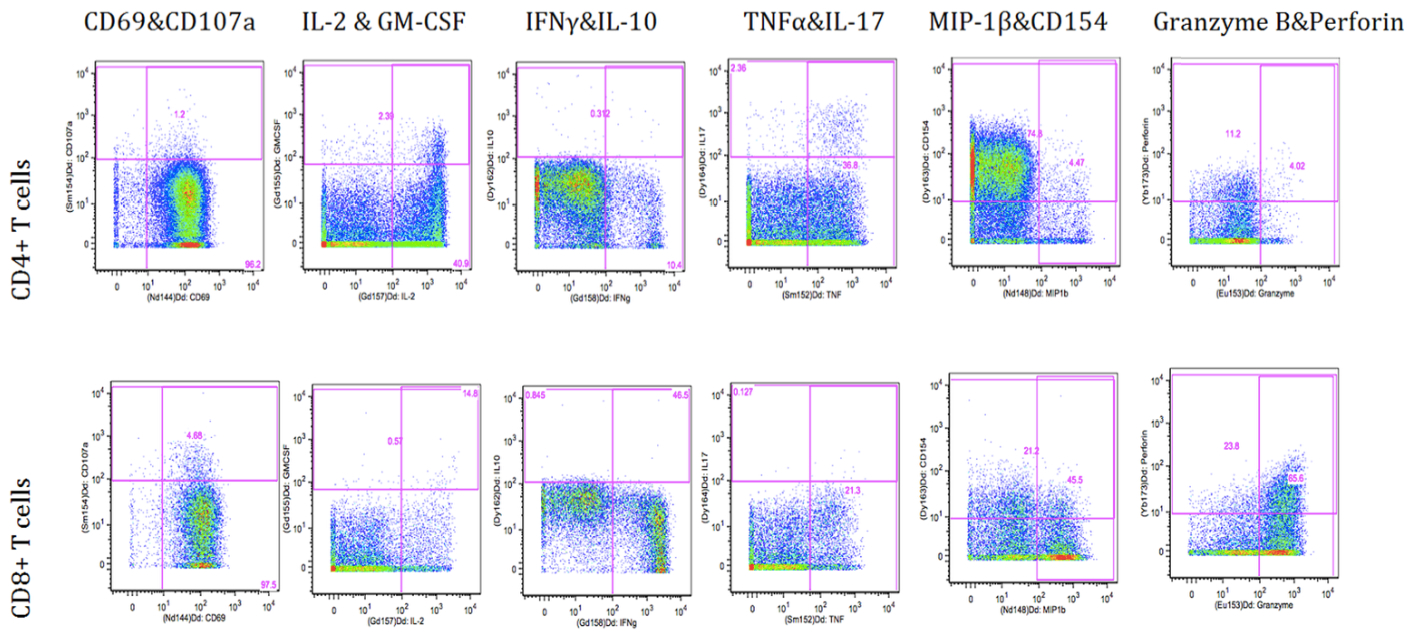
Figure 1. Flowjo gating of intracelluar markers upon PMA/Ionomycin stimulation
Recipes
- Complete RPMI
RPMI1640
10% FBS
1x pen-strep -Glutamine
- CyPBS
1x PBS without heavy metal contaminants, such as 10x PBS
Made in MilliQ water
- CyFACS buffer
1x CyPBS with 0.1% BSA
2 mM EDTA and 0.05% sodium azide
Made in MilliQ water
Note: Do not use FBS!
- Live-dead stain
5 mg/ml maleimide-DOTA loaded with 139-Lanthanum* or 115-Indium*
(*Natural-abundance metal chloride salt used; >95% specified isotope; trace-metal pure 99.99%)
Acknowledgments
This work was supported by grants # S10RR027582 and 5U19AI057229 from the National Institutes of Health. We thank Evan Newell and Mark Davis for help in the initial protocol development.
References
- Newell, E. W., Sigal, N., Bendall, S. C., Nolan, G. P. and Davis, M. M. (2012). Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36(1): 142-152.
- Bendall, S. C., Simonds, E. F., Qiu, P., Amir el, A. D., Krutzik, P. O., Finck, R., Bruggner, R. V., Melamed, R., Trejo, A., Ornatsky, O. I., Balderas, R. S., Plevritis, S. K., Sachs, K., Pe'er, D., Tanner, S. D. and Nolan, G. P. (2011). Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332(6030): 687-696.
- Maecker, H. T. (2009). Multiparameter flow cytometry monitoring of T cell responses. Methods Mol Biol 485: 375-391.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lin, D., Gupta, S. and Maecker, H. T. (2015). Intracellular Cytokine Staining on PBMCs Using CyTOFTM Mass Cytometry. Bio-protocol 5(1): e1370. DOI: 10.21769/BioProtoc.1370.
Category
Immunology > Immune cell staining > Mass cytometry
Immunology > Immune cell function > Cytokine
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









