- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Imaging and Quantitative Analysis of Size and Distribution of Spherical Bodies, e.g. Embryonic Oil Bodies
Published: Vol 5, Iss 1, Jan 5, 2015 DOI: 10.21769/BioProtoc.1369 Views: 9062
Reviewed by: Renate WeizbauerArsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views

A Simple Protocol for Periodic Live Cell Observation of Flagellate Stages in the Lichen Alga Trebouxia
Enrico Boccato [...] Mauro Tretiach
Jan 20, 2026 188 Views
Abstract
Oil bodies (OBs) are seed-specific lipid storage organelles that allow the accumulation of neutral lipids that sustain plantlet development after the onset of germination. Using fluorescent dyes and confocal microscopy, we monitored the dynamics of OBs in living Arabidopsis (Arabidopsis thaliana) embryos at different stages of development (Miquel et al., 2014). Image acquisition was followed by a detailed statistical analysis of OB size and distribution during seed development in the four dimensions (x, y, z, and t).
Keywords: Oil bodyMaterials and Reagents
- Plant material
- Arabidopsis thaliana plants, wild type or mutant or transgenic plants.
- Developing siliques between 6 and 11 days after pollination of plants grown in a greenhouse under the following conditions (13 h of light, diurnal temperature of 25 °C, and nocturnal temperature of 17 °C), and irrigated twice per week with mineral nutrient solution.
- Arabidopsis thaliana plants, wild type or mutant or transgenic plants.
- Nile Red (a neutral lipid stain, at 2 μg/ml final in acetone) (Sigma-Aldrich, catalog number: 72485 )
Equipment
- Confocal microscope
Note: In this study an inverted LEICA SP2-AOBS spectral confocal laser microscope (Leica Microsystems) equipped with an HCX PL APO CS 40 x 1.25 objective and a multiline argon laser was used. - Forceps (Dumont No.5) (Sigma-Aldrich)
- Scalpel (11 P, blade) (Swann Norton)
- Glass slides or glass-bottom dish, cover slips
- Binocular (Nikon Corporation, model: SMZ1000 ) (Champigny sur Marne with a led ring - Shott easyledTM for illumination)
Software
- ND-SAFIR (http://serpico.rennes.inria.fr/doku.php?id=software:nd-safir:index)
- AVIZO® FIRE (http://www.vsg3d.com/avizo/fire)
Procedure
- Sample preparation
- Remove developing seeds from 2-3 siliques under the binocular with a sharp scalpel and forceps (DUMONT N°5) and place them on a glass slide or in a glass-bottom dish.
- Add a small volume (between 20 and 30 µl) of Nile Red solution and cover with a cover slip. Remove embryos from the seed teguments by gently pressing seeds between the slide and cover slip with the rounded end of the handle of the scalpel and observe after 30 min of incubation in the dark at room temperature (see Video 1).Video 1. Dissecting embryos out of Arabidopsis developing siliques
Note: You can use a 60% (v/v) glycerol solution or 0.4 % (w/v) of low melting agarose as mounting medium for your staining solution to better immobilize the embryos for long observation periods.
- Remove developing seeds from 2-3 siliques under the binocular with a sharp scalpel and forceps (DUMONT N°5) and place them on a glass slide or in a glass-bottom dish.
- Confocal microscopy
- Find appropriate embryos and adjust the focus on cotyledon edges using a 40x objective.
- Set optical parameters to optimize signal to noise ratio.
- Excitation laser: the argon laser is set at 10% of 488 nm for Nile Red excitation.
- Emission band of 550 to 650 nm.
- Capture the images (90 to 100 optical sections of 0.16 µm thickness) with a 40x/1.25 oil objective and 8x digital zooming. Each section is the average of two scans conducted at the resolution of 512 x 512 pixels with a spatial resolution of 0.09 mm x 0.09 mm x 0.16 µm in the x, y, z referential.
Notes:- The Nile Red stain is highly fluorescent when associated with neutral lipids. Nevertheless you can bleach it if using too high laser power for excitation. According to our experience, reducing the laser power to its minimal value and laser excitation to 10% of laser power is far enough for excellent observation and preserves fluorescence for up to one night.
- As the fluorescence signal is usually very strong, you can decrease the pinhole’s size to 0.7 Airy to reduce the thickness of the optical section.
- Increasing scanning speed as well as using bidirectional-scanning mode will speed up acquisition.
- The Nile Red stain is highly fluorescent when associated with neutral lipids. Nevertheless you can bleach it if using too high laser power for excitation. According to our experience, reducing the laser power to its minimal value and laser excitation to 10% of laser power is far enough for excellent observation and preserves fluorescence for up to one night.
- Excitation laser: the argon laser is set at 10% of 488 nm for Nile Red excitation.
- Find appropriate embryos and adjust the focus on cotyledon edges using a 40x objective.
- Quantitative analysis
- Obtain images at a similar location from 3 different embryos for statistical analysis.
- Analyze the images using the following pipeline:
- The crude image stacks present several characteristics that disturb direct automated counting of the oil bodies (Figure 1). To circumvent this problem, the procedures for image segmentation by watershed and data extraction were implemented in AVIZO® FIRE in a semi-automated pipeline. A specific module (WshedBinSeparate) has been developed in TCL language (Tool Command Language). The outline of the procedure is described in Figure 2. A movie shows the sequence of operations (Video 2).

Figure 1. 2D/3D view of an image stack with various problems preventing direct use in image analysis. A. Images with high noise; B. Non homogenous objects in size; C. Variable grey levels; D. Objects with touching edges.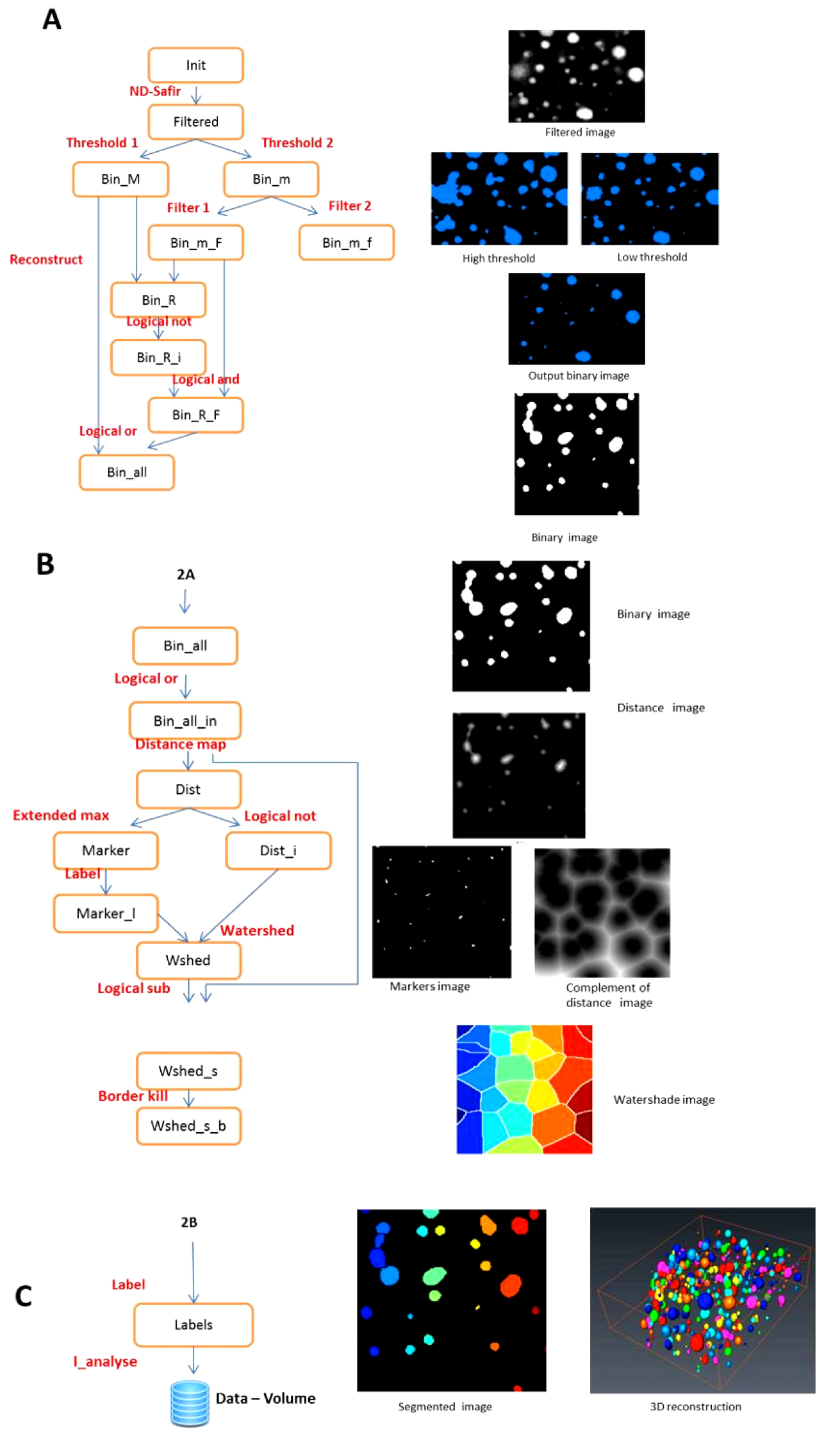
Figure 2. Image analysis pipeline. A. Generating binary images with double thresholding; B. Generating watershaded images; C. Individual labelling of objects and 3 D rendering. The segmentation chain in Avizo (left) and the resulting images (right).Video 2. OB image processing pipeline - The WshedBinSeparate module allows segmentation of oil bodies images stacks by watershed methods.
- The input image is a 3D stack in grey scale. The images are first de-noised with a Gaussian filter using ND-SAFIR. Two thresholding operations are then performed with threshold values manually indicated by the operator. The best value is chosen by visual test of various threshold values.
- The high threshold allows the detection of large objects without worrying about over- or under-estimating them but without consideration of small objects.
- The low threshold allows the detection of small objects without worrying about the over- or under-estimation of the large objects. The images are combined to produce a thresholded “binary image”.
- The 3D distance transform of this binary image is computed (a value is given for each pixel of the image according to its distance to the border of the closest object in the vicinity).
- The maximum of the distance function produces the “marker image” i.e. the centre of the segmented quasi-spherical filled object. The complement of the distance transform (CDT), with the marker image is used to create the “watershed lines”: the watershed algorithm expands the markers (like inflating a balloon) toward increasing value of the CDT. The watershed lines represent the boundaries separating the objects. Finally, the watershed lines are subtracted from the binary image to obtain separated and individualized objects (“segmented image”). The image of separated objects is then converted to a spread sheet document where each item in the spread sheet is a labeled object volume.
- The crude image stacks present several characteristics that disturb direct automated counting of the oil bodies (Figure 1). To circumvent this problem, the procedures for image segmentation by watershed and data extraction were implemented in AVIZO® FIRE in a semi-automated pipeline. A specific module (WshedBinSeparate) has been developed in TCL language (Tool Command Language). The outline of the procedure is described in Figure 2. A movie shows the sequence of operations (Video 2).
- Obtain images at a similar location from 3 different embryos for statistical analysis.
Acknowledgments
This work was funded by INRA and, in particular, by the divisions BAP (Plant Biology and Breeding) and MIA (Applied Mathematics and Informatics) for G. Trigui’s Ph.D. This work was adapted from Miquel et al. (2014).
References
- Miquel, M., Trigui, G., d'Andrea, S., Kelemen, Z., Baud, S., Berger, A., Deruyffelaere, C., Trubuil, A., Lepiniec, L. and Dubreucq, B. (2014). Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds. Plant Physiol 164(4): 1866-1878.
- Pierre, S. (2003). Morphological image analysis: Principles and Applications. 2nd edition. Springer.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Miquel, M., Trigui, G., Trubuil, A. and Dubreucq, B. (2015). Imaging and Quantitative Analysis of Size and Distribution of Spherical Bodies, e.g. Embryonic Oil Bodies. Bio-protocol 5(1): e1369. DOI: 10.21769/BioProtoc.1369.
- Miquel, M., Trigui, G., d'Andrea, S., Kelemen, Z., Baud, S., Berger, A., Deruyffelaere, C., Trubuil, A., Lepiniec, L. and Dubreucq, B. (2014). Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds. Plant Physiol 164(4): 1866-1878.
Category
Plant Science > Plant cell biology > Cell staining
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











