- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA Chromatin Immunoprecipitation (RNA-ChIP) in Caenorhabditis elegans
Published: Vol 4, Iss 24, Dec 20, 2014 DOI: 10.21769/BioProtoc.1358 Views: 15660
Reviewed by: Fanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification of Proteins Interacting with Genomic Regions of Interest in vivo Using Engineered DNA-binding Molecule-mediated Chromatin Immunoprecipitation (enChIP)
Toshitsugu Fujita and Hodaka Fujii
May 20, 2014 15467 Views

Chromatin Immunoprecipitation (ChIP) Assay for Detecting Direct and Indirect Protein – DNA Interactions in Magnaporthe oryzae
Gang Li [...] Richard A. Wilson
Nov 5, 2015 15991 Views
Abstract
The RNA chromatin immunoprecipitation assay (RNA-ChIP) allows detection and quantification of RNA–protein interactions using in vivo cross-linking with formaldehyde followed by immunoprecipitation of the RNA–protein complexes. Here we describe the RNA–ChIP protocol that we have adapted for Caenorhabditis elegans (C. elegans) to detect interaction between the nuclear Argonaute CSR-1 (chromosome segregation and RNAi deficient) protein and its target nascent RNAs. We have used a transgenic strain expressing a recombinant long isoform of CSR-1 protein fused with N-terminal 3x FLAG epitope.
Materials and Reagents
- Escherichia coli (E. coli) OP50 [available from the Caenorhabditis Genetics Center (CGC)]
- Anti–FLAG M2 affinity gel (Sigma-Aldrich, catalog number: A2220 )
- 1-Bromo-3-chloropropane (BCP) (Sigma-Aldrich, catalog number: B9673 )
- 1 M CaCl2 solution (Sigma-Aldrich, catalog number: 21114 )
- Distilled water (DNase/RNase-free UltraPureTM) (Life Technologies, InvitrogenTM, catalog number: 10977-015 )
- 10 mM dNTP mix (Thermo Fisher Scientific, catalog number: R0191 )
- EDTA (0.5 M, pH 8.0) UltraPureTM (Life Technologies, InvitrogenTM, catalog number: 15575-020 )
- Ethanol 200 proof (for molecular biology) (Sigma-Aldrich, catalog number: E7023 )
- FLAG peptide (Sigma-Aldrich, catalog number: F4799 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- GlycoBlueTM coprecipitant (15 mg/ml) (Life Technologies, Ambion®, catalog number: AM4530)
- 100x halt protease inhibitor cocktail (Thermo Fisher Scientific, catalog number: 78430 )
- Isopropanol (2-propanol, for molecular biology) (Sigma-Aldrich, catalog number: I9516 )
- KH2PO4 (Sigma-Aldrich, catalog number: P0662 )
- 8 M LiCl solution (Sigma-Aldrich, catalog number: L7026 )
- Maxima reverse transcriptase (Thermo Fisher Scientific, catalog number: EP0741 )
- 1 M MgCl2 solution (Sigma-Aldrich, catalog number: M1028 )
- 1 M MgSO4 solution (Sigma-Aldrich, catalog number: M3409 )
- Na2HPO4 (Sigma-Aldrich, catalog number: S3264 )
- 5 M NaCl solution (Life Technologies, Ambion®, catalog number: AM9759 )
- NP-40 [10% (w/v) aqueous solution] (Pierce Antibodies, catalog number: 85124 )
- Paraformaldehyde (32% solution, EM grade) (VWR International, catalog number: 15714-S )
- Phase lock gel heavy (1.5 ml) (5PRIME, catalog number: 2302810 )
- Phenol:Chloroform:IAA (25:24:1, pH 6.6) (Life Technologies, Ambion®, catalog number: AM9730 )
- Proteinase K (~20 mg/ml) (recombinant) (PCR grade) (Thermo Fisher Scientific, catalog number: EO0491 )
- 2x QuantiFast SYBR® Green qPCR master mix (QIAGEN, catalog number: 204154 )
- Random hexamers (100 µM) (Thermo Fisher Scientific, catalog number: SO142 )
- RNase inhibitor SUPERase InTM (20 U/μl) (Life Technologies, Ambion®, catalog number: AM2694 )
- 20% SDS solution (Life Technologies, Ambion®, catalog number: AM9820 )
- 3 M sodium acetate (pH 5.5) solution (Life Technologies, Ambion®, catalog number: AM9740 )
- Sodium deoxycholate stock solution [10% (w/v) in distilled water] (Pierce Antibodies, catalog number: 89904 )
- TRI Reagent® solution (Life Technologies, Ambion®, catalog number: AM9738 )
- 1 M Tris-HCl (pH 7.5) UltraPureTM (Life Technologies, InvitrogenTM, catalog number: 15567-027 )
- Triton-X 100 detergent solution (Pierce Antibodies, catalog number: 85111 )
- The sequence of primers used in Figure 1 to detect mes-4 pre-mRNA: Forward GGATACATCAATGGAGAAATGGA (spanning exon 2 and intron 3) and Reverse ACAACTCGCGTGAAATTTACTAC (spanning intron 3)
Note: DNA oligos has been synthesized by Integrated DNA Technologies (IDT).
- 1x M9 buffer (see Recipes)
- Nuclei extraction buffer (see Recipes)
- RIPA buffer (see Recipes)
- TSE 150 buffer (see Recipes)
- TSE 500 buffer (see Recipes)
- TSE 1,000 buffer (see Recipes)
- LiCl buffer (see Recipes)
- TE buffer (see Recipes)
- Elution buffer (see Recipes)
Equipment
- 1.5 ml Bioruptor TPX® polymethylpentene tubes (Diagenode, catalog number: M500-50 )
- Microfuge tubes (1.5 ml, RNase free) (Life Technologies, InvitrogenTM, catalog number: AM12400 )
- Bioruptor® standard UCD-200 sonicator (Diagenode, catalog number: B01010003 )
- Dura-GrindTM stainless steel dounce tissue grinder 7 ml size (Wheaton Scientific, catalog number: 357572 )
- Real-time PCR machine (e.g. Eppendorf, Mastercycler® ep realplex4)
- Refrigerated tabletop microcentrifuge
- LabquakeTM tube rotators (Thermo Fisher Scientific, catalog number: 400220Q )
- Vortex
Procedure
- Fixation and nuclei extraction
- Grow synchronized populations of worms on one large LB agar plate (100 x 15 mm) seeded with OP50 bacteria (~20,000 worms).
- Wash worms off the plate with cold M9.
- Centrifuge for 1 min at 1,200 x g to pellet the worms.
- Wash 3 times with cold M9.
- Centrifuge for 1 min at 1,200 x g to pellet the worms.
- Fix the worms with 2% paraformaldehyde in 10 ml M9 buffer for 30 min at room temperature on rotation.
- Centrifuge for 1 min at 1,200 x g to pellet the worms.
- Discard the supernatant and wash one time with 10 ml cold 0.1 M Tris-HCl (pH 7.5).
- Centrifuge for 1 min at 1,200 x g to pellet the worms.
- Wash the worm pellet twice with 10 ml cold M9 buffers.
- Centrifuge for 1 min at 1,200 x g to pellet the worms and discard the supernatant.
Note: At this step you can freeze the worm pellet and store at -80 °C.
- Resuspend worm pellet in 2 ml cold Nuclei extraction buffer.
- Transfer 2 ml of worms to a steel dounce (on ice) and stroke at least 30 times.
Note: Check under the microscope that the worms have been crushed.
- Transfer 2 ml of lysate to two RNase free 1.5 ml microfuge tubes and centrifuge 2 min at 40 x g at 4 °C.
- Transfer the supernatant to two new RNase free 1.5 ml microfuge tubes.
- Centrifuge for 5 min at 1,000 x g at 4 °C to pellet the nuclear fraction.
- Discard the supernatant and wash the nuclear pellet with 1 ml of cold nuclei extraction buffer.
Note: Combine the two pellets in one RNase free 1.5 ml microfuge tube.
- Centrifuge for 5 min at 1,000 x g at 4 °C to pellet the nuclear fraction.
- Repeat steps 17 and 18 twice.
- Centrifuge for 5 min at 1,000 x g at 4 °C to pellet the nuclear fraction.
- Resuspend the nuclear pellet in 500 μl RIPA buffer.
- Grow synchronized populations of worms on one large LB agar plate (100 x 15 mm) seeded with OP50 bacteria (~20,000 worms).
- Sonication
- Transfer the lysate to a 1.5 ml Bioruptor TPX® polymethylpentene tube.
- Sonicate sample for 20 min at 4 °C (settings: 30 sec on, 30 sec off, level high 320 W, wave frequency 20 kHz).
- Centrifuge for 5 min at 12,000 x g at 4 °C. Transfer the supernatant (containing chromatin) to a new RNase free 1.5 ml microfuge tube.
Note: At this step samples can be stored at -80 °C.
- Transfer the lysate to a 1.5 ml Bioruptor TPX® polymethylpentene tube.
- Immunoprecipitation
- Take 1/20 fraction of the supernatant for the input RNA extraction and store on ice.
- Use 30 μl of anti-Flag M2 affinity gel for each immunoprecipitation.
- Equilibrate the affinity gel with 1 ml of RIPA buffer.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Repeat steps 27 and 28.
- Add supernatant from step 24 to the equilibrated affinity gel.
- Incubate on rotation (continuous rotation at 8rpm) for 1 h at 4 °C.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Wash the affinity gel with 1 ml TSE150 buffer and incubate on rotation for 1 min at 4 °C.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Discard the supernatant and wash the affinity gel 2 times with 1 ml TSE500 buffer and incubate on rotation for 1 min at 4 °C.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Discard the supernatant and wash the affinity gel with 1 ml TSE1000 buffer and incubate on rotation for 5 min at 4 °C.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Discard the supernatant and wash the affinity gel with 1 ml LiCl buffer and incubate on rotation for 5 min at 4 °C.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Discard the supernatant and wash the affinity gel 2 times with 1 ml TE buffer.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Discard the supernatant and remove as much as possible buffer using P200 pipette tips.
- Elute the protein-RNA complexes from the affinity gel using 200 μl of elution buffer.
- Incubate with rotation for 1 h at 4 °C.
- Centrifuge for 1 min at 1,000 x g at 4 °C.
- Transfer the supernatant to a new RNase free 1.5 ml microfuge tube.
- Take 1/20 fraction of the supernatant for the input RNA extraction and store on ice.
- Reverse crosslink
- Take the 200 μl of supernatant from step 47 and input RNA from step 25 (to a final volume of 200 μl with elution buffer) and treat them with 1 μl (20 μg) of proteinase K for 1 h at 45 °C.
- RNA purification
- Add 750 μl of TRI Reagent® solution to the samples from step 48 and vortex for 30 sec.
- Add 100 μl of BCP and vortex for 30 sec.
- Incubate at room temperature for 10 min.
- Centrifuge for 15 min at 12,000 x g at 4 °C.
- Transfer the top aqueous phase in a new RNase free 1.5 ml microfuge tube.
- Add 1.5 μl of GlycoblueTM and 50 μl of 3 M sodium acetate.
- Mix and add 1 volume of isopropanol (typically 400-500 μl).
- Incubate at room temperature for 5 min.
- Centrifuge for 20 min at 12,000 x g at 4 °C.
- Wash the RNA pellet with 1 ml of 70% ethanol.
- Centrifuge for 5 min at 7,500 x g at 4 °C.
- Resuspend the pellet in 44 μl of RNase-free UltraPureTM distilled water.
- Add 750 μl of TRI Reagent® solution to the samples from step 48 and vortex for 30 sec.
- DNase treatment
- Add 5 μl of 10x TURBO DNase buffer and 1 μl of TURBO DNase to the samples from step 60.
- Incubate 30 min at 37 °C.
- Add 450 μl of RNase-Free UltraPureTM distilled water (total volume 500 μl).
- Add 500 μl of Phenol:Chloroform:IAA and mix by vortex.
- Transfer the mixture to 1.5 ml Phase Lock Gel tube and centrifuge at 12,000 x g for 5 min at 4 °C.
- Transfer top aqueous phase in a new RNase free 1.5 ml microfuge tube.
- Add 1.5 μl of GlycoblueTM and 50 μl of 3 M sodium acetate and mix by vortex.
- Add 1 volume of isopropanol (typically 400-500 μl) and mix by vortex.
- Incubate at room temperature for 5 min.
- Centrifuge for 20 min at 12,000 x g at 4 °C.
- Wash the RNA pellet with 1 ml of 70% ethanol.
- Centrifuge for 5 min at 7,500 x g at 4 °C.
- Resuspend in 12.5 μl ofRNase-Free UltraPureTM distilled water.
- Add 5 μl of 10x TURBO DNase buffer and 1 μl of TURBO DNase to the samples from step 60.
- cDNA Synthesis
- Add to the samples from step 73, 1 μl of random hexamer primers, 4 μl of 5x reaction buffer, 0.5 μl of RNase Inhibitor, 1 μl of dNTP mix (0.5 mM final concentration), and 1 μl of Maxima reverse transcriptase.
- Incubate 10 min at 25 °C followed by 60 min at 37 °C.
- Stop the reaction by heating at 70 °C for 10 min.
Note: The reverse transcription reaction can be stored at -20 °C.
- Add to the samples from step 73, 1 μl of random hexamer primers, 4 μl of 5x reaction buffer, 0.5 μl of RNase Inhibitor, 1 μl of dNTP mix (0.5 mM final concentration), and 1 μl of Maxima reverse transcriptase.
- qPCR
- Dilute the 20 μl sample from step 76 with 100 μl of nuclease free water and use 5 μl as a template in 25 μl qPCR reaction (performed in triplicate reactions); add 12.5 μl of 2x QuantiFast SYBR® Green mix, and primers to a final concentration of 0.6 μM. For the amplification of nascent RNAs, the primers have to be designed in such a way that at least one PCR primer (forward or reverse) is located in an intron of a target gene and the amplicon is 70-100 bp long. The average Ct value from triplicate qPCR reactions will be consider to calculate the percentage of input. For more information about the percentage of input calculation see http://www.lifetechnologies.com/us/en/home/life-science/epigenetics-noncoding-rna-research/chromatin-remodeling/chromatin-immunoprecipitation-chip/chip-analysis.html.
- For qPCR cycling conditions use the parameters suggested by QuantiFast SYBR® Green mix manufacturers.
Note: The best way to represent the data is by percentage of input (see Figure 1).
- Dilute the 20 μl sample from step 76 with 100 μl of nuclease free water and use 5 μl as a template in 25 μl qPCR reaction (performed in triplicate reactions); add 12.5 μl of 2x QuantiFast SYBR® Green mix, and primers to a final concentration of 0.6 μM. For the amplification of nascent RNAs, the primers have to be designed in such a way that at least one PCR primer (forward or reverse) is located in an intron of a target gene and the amplicon is 70-100 bp long. The average Ct value from triplicate qPCR reactions will be consider to calculate the percentage of input. For more information about the percentage of input calculation see http://www.lifetechnologies.com/us/en/home/life-science/epigenetics-noncoding-rna-research/chromatin-remodeling/chromatin-immunoprecipitation-chip/chip-analysis.html.
Representative data
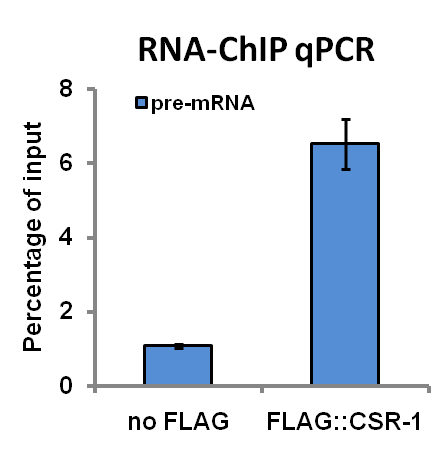
Figure 1. RNA–ChIP qPCR results obtained with an antibody against FLAG epitope and nuclear extracts from transgenic worms expressing FLAG:CSR-1 protein and control non-transgenic worms (no FLAG). The primers used in a qPCR assay have been designed to span one intron of the pre-mRNA of a known target transcript of CSR-1. The results from three independent experiments (biological replicates) are shown as percentage of input. Error bars represent the standard deviation.
Notes
The protocol has been applied to the transgenic strain expressing a recombinant CSR-1 protein fused with Flag epitope. However, it can be used with virtually any Flag-tagged nuclear proteins expressed in C. elegans. Moreover, if a primary antibody raised against the protein of interest is available, it can be crosslinked to Protein A/G resin and used instead of the anti-Flag M2 affinity gel. In this case, it is recommended to choose an antibody that has been tested in ChIP experiments. Because of the high background of the RNA-ChIP assay, it is highly recommended to always use a non-transgenic strain as a control in each experiment or knock out strains if primary antibody raised against the protein of interest will be used. The following protocol can also be used for cytoplasmic RNA-binding proteins. In this case, the nuclear extraction step can be omitted.
Recipes
- 1x M9 buffer
3 g KH2PO4
6 g Na2HPO4
17 ml 5 M NaCl
1 ml 1 M MgSO4 in 1 L of deionized water
- Nuclei extraction buffer
10 mM Tris-HCl (pH 7.5)
2 mM MgCl2
3 mM CaCl2
0.5% NP-40
10% glycerol
100 U/ml RNase inhibitor
1x protease inhibitor cocktails
- RIPA buffer
0.1% SDS
0.1% deoxycholate
1% Triton X-100
1 mM EDTA
10 mM Tris-HCl (pH 7.5)
150 mM NaCl
100 U/ml RNase inhibitor, protease inhibitor cocktails
- TSE 150 buffer
0.1% SDS
1% Triton X-100
2 mM EDTA
20 mM Tris-HCl (pH 7.5)
150 mM NaCl
100 U/ml RNase inhibitor
- TSE 500 buffer
Same as TSE 150 but with 500 mM NaCl
- TSE 1,000 buffer
Same as TSE 150 but with 1 M NaCl
- LiCl buffer
250 mM LiCl
1% NP40
1% deoxycholate
1 mM EDTA
10 mM Tris-HCl (pH 7.5)
100 U/ml RNase inhibitor
- TE buffer
10 mM Tris-HCl (pH 7.5)
1 mM EDTA
- Elution buffer
10 mM Tris-HCl (pH 7.5)
0.5% NP-40
200 mM NaCl
300 μg/ml FLAG peptide
100 U/ml RNase inhibitor
Acknowledgments
The transgenic strain expressing a recombinant CSR-1 protein fused with 3x FLAG epitope was kindly provided by C. Mello (University of Massachusetts, Worcester). This work was supported by US National Institutes of Health grant 1DP2OD006412-01 (A. G.). This protocol has been adapted from our previous work (Cecere et al., 2014).
References
- Bittencourt, D. and Auboeuf, D. (2012). Analysis of co-transcriptional RNA processing by RNA-ChIP assay. Methods Mol Biol 809: 563-577.
- Cecere, G., Hoersch, S., O'Keeffe, S., Sachidanandam, R. and Grishok, A. (2014). Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat Struct Mol Biol 21(4): 358-365.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cecere, G. and Grishok, A. (2014). RNA Chromatin Immunoprecipitation (RNA-ChIP) in Caenorhabditis elegans. Bio-protocol 4(24): e1358. DOI: 10.21769/BioProtoc.1358.
Category
Biochemistry > Protein > Immunodetection > ChIP
Biochemistry > RNA > RNA-protein interaction
Molecular Biology > RNA > RNA-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











