- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
TGFβ Release Co-culture Assay
Published: Vol 4, Iss 23, Dec 5, 2014 DOI: 10.21769/BioProtoc.1314 Views: 11977
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection of Wnt5 in Media Conditioned by Mouse Embryonic Fibroblast
Juan Flores and Nan Gao
Oct 20, 2016 8994 Views

Quantitative Live-cell Reporter Assay for Noncanonical Wnt Activity
Edith P. Karuna [...] Hsin-Yi Henry Ho
Mar 20, 2018 9837 Views

Organotypic Slice Culture of the Embryonic Mouse Brain
James M. Clegg and Thomas Pratt
Jul 5, 2020 5892 Views
Abstract
TGFβ is a potent cytokine modulating various processes including proliferation, differentiation, ECM synthesis and apoptosis (Siegel and Massague, 2003). Thus in many tissues availability of TGFβ is tightly regulated. TGFβ is secreted as an inactive complex where it is encapsulated by the latency associated protein (LAP), a ligand trap protein, which inhibits TGFβ binding to its receptor and retains TGFβ in the extracellular matrix (ten Dijke and Arthur, 2007). TGFβ can be released from the matrix and converted into its biological active form by huge number of processes including heat, high and low pH, release of reactive oxygen species (ROS) or various proteases (e.g. plasmin, elastase, matrix metalloproteinase-2 and -9) (Barcellos-Hoff and Dix, 1996; Lyons et al., 1988; Taipale et al., 1994; Yu and Stamenkovic, 2000). However, under physiological conditions the interaction of αv-class integrins with the RGD tripeptide motif in the LAP protein represents the key factor for TGFβ release in vivo. The relevance of integrin mediated TGFβ release for in vivo development and homeostasis is further underlined by the observation that mice with the integrin-binding deficient LAP proteins (RGD motif mutated to RGE) recapitulate all major phenotypes of TGFβ1 null mice, including multi-organ inflammation and defects in vasculogenesis (Shull et al., 1992; Yang et al., 2007). This striking phenotype overlap with TGFβ deficient mice and phenotypes of mice lacking αv-class integrins (Aluwihare et al., 2009; Bader et al., 1998) demonstrates an essential interconnection of integrins with TGFβ signaling in vivo, while the role of non-integrin mediated release mechanisms (ROS, pH, proteolytic cleavage etc.) during development remains less clear.
The TGFβ release assay measures the ability of cells to release TGFβ from a matrix. The assay was developed by (Annes et al., 2004) and we further optimized the protocol for keratinocytes. For other cell types the cell culture medium and culturing conditions would need to be adapted accordingly.
In keratinocytes TGFβ release is mainly mediated by αvβ6 integrin but also integrin αvβ3, αvβ5 and αvβ8 have been shown to liberate TGFβ, while other RGD binding integrins, such as α5β1 or α8β1 cannot release TGFβ (Asano et al., 2005a, 2005b; Mu et al., 2002; Munger et al., 1999). Mechanistically, the interaction with αvβ3, αvβ5 or αvβ6 integrin induces a conformational change in the LAP-TGFβ by generating an actin cytoskeleton dependent pulling force, allowing TGFβ to access its receptors. For αvβ8 integrin mediated TGFβ release it was shown that proteolytic cleavage is involved [see (Mu et al., 2002) for blocking conditions of TGFβ release by proteolytic cleavage and αvβ8 integrin].
The following protocol is optimized for the study of αvβ6-integrin mediated TGFβ release in keratinocytes.
Materials and Reagents
- Cell lines
- CHO-LTBP1 TGFβ rich matrix producing cell line, generated by the Daniel Rifkin lab expresses high levels of LTBP1-TGFβ (Annes et al., 2004).
- Transformed mink lung epithelial cells (tMLEC) TGFβ reporter cell line, stably expresses a luciferase reporter plasmid under control of a truncated plasminogen activator inhibitor type 1 promoter (PAI-1) (Abe et al., 1998).
Note: CHO-LTBP1 and tMLEC are cultured in DMEM growth medium. tMLEC DMEM growth medium is supplemented with 250 mg/ml Geneticin (Life Technologies, InvitrogenTM, catalog number: 10131035 ).
- CHO-LTBP1 TGFβ rich matrix producing cell line, generated by the Daniel Rifkin lab expresses high levels of LTBP1-TGFβ (Annes et al., 2004).
- Antibodies
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E6758 )
- BrightGlo luciferase assay kit (Promega Corporation, catalog number: E2610 )
- Dulbecco's minimum essential medium (DMEM) (Life Technologies, Gibco®, catalog number: 11966-025 )
- Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10270-106 )
- Calcium chloride (CaCl2) (Carl Roth, catalog number: A119.1 )
- Chelex 100 resin (Bio-Rad Laboratories, catalog number: 143-2832 )
- Penicillin-streptomycin (pen-strep) (Life Technologies, Gibco®, catalog number: 15070-063 )
- Minimum essential medium (MEM) (Sigma-Aldrich, catalog number: M8167 )
- Insulin (Sigma-Aldrich, catalog number: I5500 )
- Epidermal Growth Factor (EGF) (Sigma-Aldrich, catalog number: E9644 )
- Transferin (Sigma-Aldrich, catalog number: T8158 )
- Phosphoethanolamine (Sigma-Aldrich, catalog number: P0503 )
- Ethanolamine (Sigma-Aldrich, catalog number: E0135 )
- Hydrocortisone (Calbiochem®, catalog number: 386698 )
- L-Glutamine (Life Technologies, InvitrogenTM, catalog number: 25030-081 )
- Trypsin powder (Life Technologies, Gibco®, catalog number: 27250-018 )
- 0.5%Trypsin/EDTA (Life Technologies, Gibco®, catalog number: 15400-054 )
- Sodium chloride (NaCl) (Carl Roth, catalog number: P029 )
- di-Sodium hydrogen phosphate (Na2HPO4) (Carl Roth, catalog number: T876 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- Potassium chloride (KCl) (Carl Roth, catalog number: HN02.3 )
- Potassium dihydrogen phosphate (KH2PO4) (Carl Roth, catalog number: 3904 )
- DMEM growth medium (see Recipes)
- Keratinocyte growth medium for murine keratinocyte culture (KGM) (see Recipes)
- Starving KGM (see Recipes)
- 0.4% keratinocyte trypsin (see Recipes)
- Phosphate-buffered saline (PBS) (see Recipes)
- PBS/EDTA (see Recipes)
- 0.1% trypsin/EDTA (see Recipes)
- Chelated FBS (see Recipes)
Equipment
- Flat bottom 96-well plates for cell culture (Corning, Costar®, catalog number: 3595 )
- Round bottom white 96-well plates (Corning, Costar®, catalog number: 3789 )
- 1.5 ml Eppendorf tubes (Sigma-Aldrich, catalog number: T9661-1000EA )
- Cell culture incubator (37 °C, 5% CO2)
- Standard centrifuge to spin down the cells
- Luminometer, e.g. GloMax (Promega corporation)
- Standard bright field microscope
- Multichannel pipette (recommended)
Procedure
- Considerations before you get started
- Consider carefully how many wells you need to coat with a TGFβ rich-matrix in order to perform all necessary control conditions, since matrix generation is the most time consuming step in this protocol. It is further recommended to prepare more wells, because it can happen at later steps that the matrix gets lost or CHO-LTBP1 cells cannot be removed properly in single wells.
- Cells which have been thawed should be passaged twice prior to the experiment.
- Each condition should be analyzed in triplicates.
- Conditions to include
- Only tMLEC → reporter cell back ground signal
- Cell of interest + tMLEC → untreated sample
- Cell of interest + tMLEC + αvβ6 blocking antibody → αvβ6 dependent TGFβ release control
- Cell of interest + tMLEC + TGFβ neutralizing antibody → TGFβ dependent luciferase reporter control. (This antibody blocks TGFβ binding to its high affinity TGFβ receptor.)
→ Condition c can be extended or modified according to the TGFβ release mechanism which will be analyzed; e.g. for proteolytic and αvβ8 integrin-mediated release, see (Mu et al., 2002).
- Only tMLEC → reporter cell back ground signal
- During the assay there is no way to check for matrix loss as the matrix is difficult to see with bright field microscope. In parallel wells could be prepared where after cell removal the matrix is stained with antibodies against matrix components such as fibronectin and LTBP proteins.
- Consider carefully how many wells you need to coat with a TGFβ rich-matrix in order to perform all necessary control conditions, since matrix generation is the most time consuming step in this protocol. It is further recommended to prepare more wells, because it can happen at later steps that the matrix gets lost or CHO-LTBP1 cells cannot be removed properly in single wells.
- Preparation of a cell-free, TGFβ-rich matrix
- Plate 5.0 x 104 CHO-LTBP1 cells per well of a flat bottom 96-well plate in 100 µl DMEM growth medium and incubate in a cell culture incubator (37 °C, 5% CO2) for 48 h.
Note: After 48 h the plated 96-well should be completely confluent, which can be checked under a standard bright field microscope. In the next steps, it is critical to remove the CHO-LTBP1 cells without destroying the TGFβ rich matrix and keeping the plate sterile. Thus each pipetting step should be performed very carefully without scratching the bottom of the wells. To avoid drying out of the wells the use of a multichannel pipette is recommended).
- Remove medium and add 100 µl PBS/EDTA solution on top of CHO-LTBP1 cells to wash off residual medium.
- Following the washing step add 100 µl PBS/EDTA and incubate for at least 30 min at 37 °C, 5% CO2.
Note: Check with the microscope that cells round up and detach. If this is not the case prolong incubation for another 30 min.
- Remove the supernatant carefully with the multichannel pipette and add 100 µl PBS/EDTA. Gently pipette up and down 3-4 times and remove the supernatant. Repeat this step if necessary with 100 µl PBS/EDTA solution.
Note: Check each well with the microscope in between washes to ensure complete removal of cells in all wells while avoiding too vigorous washing. If not all the cells are removed, remaining CHO-LTBP1 cells will re-adhere and generate/release TGFβ, contributing to the final result and thus generating false positive signals. On the other hand too harsh washing leads to a loss of the matrix. Further do not directly pipette on top of the matrix to avoid matrix damages. Thus it is recommended to tilt the plate by 45 °C during rinsing, so that the PBS/EDTA solution gently flows over the well bottom. Nevertheless, too many repeats of this gentle rinsing process can also cause matrix detachment.
- Then wash the matrix two times with PBS only at room temperature to remove all traces of EDTA.
Note: Minimal traces of EDTA will impair keratinocyte adhesion and function! Thus this washing step is very important if you work with keratinocytes!
- Overlay the matrix coated wells with 50 µl starving KGM and keep at room temperature, while you prepare the cells, to avoid the drying of the matrix.
- Plate 5.0 x 104 CHO-LTBP1 cells per well of a flat bottom 96-well plate in 100 µl DMEM growth medium and incubate in a cell culture incubator (37 °C, 5% CO2) for 48 h.
- Co-culture of the reporter cell line and cells of interest
- Detach cells of interest and TGFβ reporter cell line (tMLEC) from cell culture dishes. (For keratinocytes use keratinocyte trypsin, otherwise Trypsin/EDTA can be used). For trypsin digestion incubate cells with trypsin solution (37 °C, 5% CO2) and as soon as cells detach and rounded up stop the digestion and collect cells by adding complete medium. Then spin down single cell suspension (5 min, 900 rpm, 78 x g). It is important to wash suspended tMLEC cells once with starving KGM, to remove Ca2+ from the original DMEM medium.
- Per well mix 2.0 x 104 cells of interest (keratinocytes) and 1.5 x 104 tMLEC in 100 µl Starving KGM (total volume) in a 1.5 ml Eppendorf tube.
- For the antibody blocking controls (condition c and d) incubate mixed cells with αvβ6 integrin–blocking antibody (20 μg/ml) or TGFβ neutralizing antibody (15 μg/ml) in the 1.5 ml Eppendorf tube for 15 min at room temperature before plating.
- Remove starving KGM from matrix coated wells and quickly plate all conditions (100 µl total volume).
- Incubate 96-well plate for 16-24 h in a cell culture incubator (37 °C, 5% CO2).
- Detach cells of interest and TGFβ reporter cell line (tMLEC) from cell culture dishes. (For keratinocytes use keratinocyte trypsin, otherwise Trypsin/EDTA can be used). For trypsin digestion incubate cells with trypsin solution (37 °C, 5% CO2) and as soon as cells detach and rounded up stop the digestion and collect cells by adding complete medium. Then spin down single cell suspension (5 min, 900 rpm, 78 x g). It is important to wash suspended tMLEC cells once with starving KGM, to remove Ca2+ from the original DMEM medium.
- Detection of luciferase activity and data analysis
Note: Procedure mainly follows the manufacturers’ protocol which can be downloaded on the Promega web site.
- Equilibrate BrightGlo reagent to room temperature and add 100 µl to each well after equilibrating the cell containing 96-well plate to room temperature for 5 min.
- Then pipet up and down 2-3 times to ensure efficient cell lysis, while avoiding air bubbles and transfer the complete content of each well (200 µl) to the white round bottom 96-well plate. (Before transfer efficient cell lysis can be quickly control under a bright field microscope.)
- Incubate for 5 min at room temperature before measuring luciferase activity with a luminometer, e.g. GloMax.
Note: Before measuring the luciferase activity, make sure that all air bubbles have been removed, as they obscure signal detection.
- For data analysis average the triplicate measurements for each well and subtract the back ground signal (tMLEC only control).
- Equilibrate BrightGlo reagent to room temperature and add 100 µl to each well after equilibrating the cell containing 96-well plate to room temperature for 5 min.
Representative data
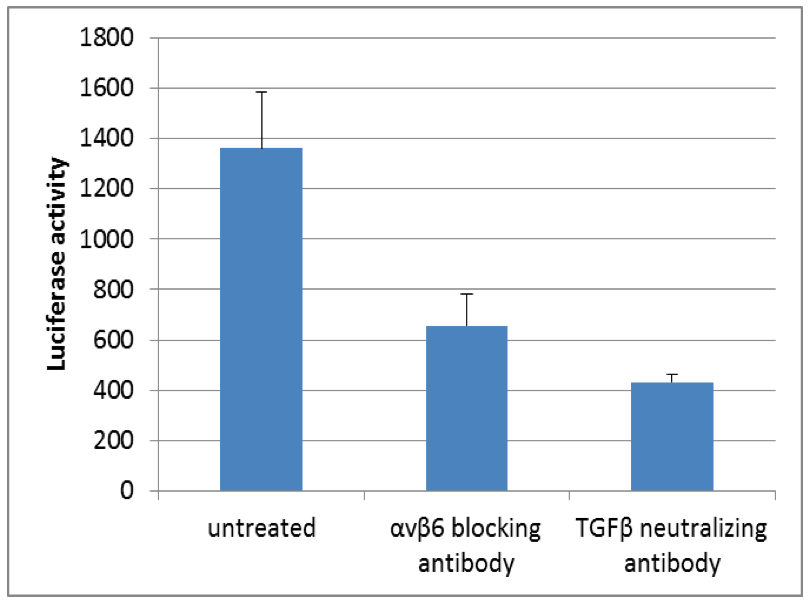
Figure 1. Luciferase activity of TGFβ reporter cell line (tMLEC) in TGFβ release co-culture assay with murine kerationcytes. Note the decrease in luciferase activity when cells are incubated with αvβ6 blocking antibody or TGFβ neutralizing antibody.
Recipes
- DMEM growth medium
Dulbecco's minimum essential medium (DMEM) containing 10% heat-inactivated FBS and 1x pen-strep
- Keratinocyte growth medium for murine keratinocyte culture (KGM)
Filter the mixture through 0.2 µm and stored at 4 °C for up to 1 monthFinal working concentration
Initial stock concentration
Vol.
MEM
500 ml
5 µg/ml insulin
5 mg/ml in 4 mM HCl
0.5 ml
10 ng/ml EGF
200 µg/ml in PBS
25 µl
10 µg/ml transferin
5 mg/ml in PBS
1 ml
10 µM phosphoethanolamine
10 mM in PBS
0.5 ml
10 µM ethanolamine
10 mM in PBS
0.5 ml
0.36 µg/ml hydrocortisone
5 mg/ml in ethanol
36 µl
1x glutamine
100x
5 ml
1x pen-strep
100x
5 ml
8% chelated FBS (Ca2+ free)
(see Recipes)
40 ml
45 µM CaCl2 (sterile filtrated)
100 mM
225 µl
- Starving KGM
Filter the mixture through 0.2 µm and stored at 4 °C for up to 1 monthFinal working concentration
Initial stock concentration
Vol.
MEM
500 ml
1x pen-strep
100x
5 ml
45 µM CaCl2 (sterile filtrated)
100 mM
225 µl
- 0.4% keratinocyte trypsin
Dissolve 0.4 g trypsin powder in 100 ml PBS and pass through a 0.2 µm filter for sterilization
It can be stored either at -20 °C for 1 year or at +4 °C for 1 month
Avoid repeated freeze thaw cycles
- Phosphate-buffered saline (PBS)
Dissolve NaCl (137 mM) 8 g/L, KCl (2.7 mM) 0.2 g/L, Na2HPO4 (10 mM) 1.44 g/L, KH2PO4 (1.8 mM) 0.24 g/L in ddH2O and adjust pH to 7.4 with HCl
- PBS/EDTA
PBS supplemented with 15 mM EDTA (should be sterile)
- Trypsin/EDTA
Diluted 0.5% trypsin/EDTA with PBS to 0.1%
- Chelated FBS
Add hydrated Chelex 100 resin to FBS (20 g of resin for 40 ml of FBS) and stir for 1 h at 4 °C
Further remove the resin by filtration and stored chelated FBS at -20 °C
Acknowledgments
The protocol is adapted from published work (Annes et al., 2004) and was funded by the Max Planck Society.
References
- Abe, M., Oda, N. and Sato, Y. (1998). Cell-associated activation of latent transforming growth factor-beta by calpain. J Cell Physiol 174(2): 186-193.
- Aluwihare, P., Mu, Z., Zhao, Z., Yu, D., Weinreb, P. H., Horan, G. S., Violette, S. M. and Munger, J. S. (2009). Mice that lack activity of alphavbeta6- and alphavbeta8-integrins reproduce the abnormalities of Tgfb1- and Tgfb3-null mice. J Cell Sci 122(Pt 2): 227-232.
- Annes, J. P., Chen, Y., Munger, J. S. and Rifkin, D. B. (2004). Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol 165(5): 723-734.
- Asano, Y., Ihn, H., Yamane, K., Jinnin, M., Mimura, Y. and Tamaki, K. (2005a). Increased expression of integrin alpha(v)beta3 contributes to the establishment of autocrine TGF-beta signaling in scleroderma fibroblasts. J Immunol 175(11): 7708-7718.
- Asano, Y., Ihn, H., Yamane, K., Jinnin, M., Mimura, Y. and Tamaki, K. (2005). Involvement of alphavbeta5 integrin-mediated activation of latent transforming growth factor beta1 in autocrine transforming growth factor beta signaling in systemic sclerosis fibroblasts. Arthritis Rheum 52(9): 2897-2905.
- Bader, B. L., Rayburn, H., Crowley, D. and Hynes, R. O. (1998). Extensive vasculogenesis, angiogenesis, and organogenesis precede lethality in mice lacking all alpha v integrins. Cell 95(4): 507-519.
- Barcellos-Hoff, M. H. and Dix, T. A. (1996). Redox-mediated activation of latent transforming growth factor-beta 1. Mol Endocrinol 10(9): 1077-1083.
- Lyons, R. M., Keski-Oja, J. and Moses, H. L. (1988). Proteolytic activation of latent transforming growth factor-beta from fibroblast-conditioned medium. J Cell Biol 106(5): 1659-1665.
- Mu, D., Cambier, S., Fjellbirkeland, L., Baron, J. L., Munger, J. S., Kawakatsu, H., Sheppard, D., Broaddus, V. C. and Nishimura, S. L. (2002). The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol 157(3): 493-507.
- Munger, J. S., Huang, X., Kawakatsu, H., Griffiths, M. J., Dalton, S. L., Wu, J., Pittet, J. F., Kaminski, N., Garat, C., Matthay, M. A., Rifkin, D. B. and Sheppard, D. (1999). The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96(3): 319-328.
- Shull, M. M., Ormsby, I., Kier, A. B., Pawlowski, S., Diebold, R. J., Yin, M., Allen, R., Sidman, C., Proetzel, G., Calvin, D. and et al. (1992). Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature 359(6397): 693-699.
- Taipale, J., Miyazono, K., Heldin, C. H. and Keski-Oja, J. (1994). Latent transforming growth factor-beta 1 associates to fibroblast extracellular matrix via latent TGF-beta binding protein. J Cell Biol 124(1-2): 171-181.
- ten Dijke, P. and Arthur, H. M. (2007). Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol 8(11): 857-869.
- Yang, Z., Mu, Z., Dabovic, B., Jurukovski, V., Yu, D., Sung, J., Xiong, X. and Munger, J. S. (2007). Absence of integrin-mediated TGFbeta1 activation in vivo recapitulates the phenotype of TGFbeta1-null mice. J Cell Biol 176(6): 787-793.
- Yu, Q. and Stamenkovic, I. (2000). Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev 14(2): 163-176.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rognoni, E. (2014). TGFβ Release Co-culture Assay. Bio-protocol 4(23): e1314. DOI: 10.21769/BioProtoc.1314.
Category
Cell Biology > Cell signaling > Development
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









