- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of ALT Associated Promyelocytic Leukemia Nuclear Bodies (APBs) by Immunofluorescence-FISH (IF-FISH)
Published: Vol 4, Iss 23, Dec 5, 2014 DOI: 10.21769/BioProtoc.1303 Views: 14454
Reviewed by: HongLok LungAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Unbiased Screening of Activated Receptor Tyrosine Kinases (RTKs) in Tumor Extracts Using a Mouse Phospho-RTK Array Kit
Julian Naipauer [...] Enrique A. Mesri
Apr 20, 2019 5680 Views

SIRF: A Single-cell Assay for in situ Protein Interaction with Nascent DNA Replication Forks
Sunetra Roy and Katharina Schlacher
Sep 20, 2019 8072 Views
Abstract
The activation of functions that counteract the physiological shortening of telomeres in rapidly proliferating cell is prerequisite for the progression of cancer cells to full malignancy (Collado et al., 2007). In most human cancers, the length of telomere is maintained through up-regulation of telomerase whereas a telomerase-independent pathway, termed Alternative Lengthening of Telomeres (ALT) is active in about 10-15% of cancers (Johnson and Broccoli, 2007; Heaphy et al., 2011). One characteristic feature of ALT is the formation of ALT-associated Promyelocytic Leukemia nuclear bodies (APBs) (Lang et al., 2010; Yeager et al., 1999). APBs contain Promyelocytic Leukemia nuclear bodies (PML-NB) components such as PML, SP100 and SUMO, telomeric DNA and telomere associated proteins including the shelterin components TRF1, TRF2, POT1, TIN2, TPP1 and Rap1 (Yeager et al., 1999). In addition, APBs contain proteins involved in DNA repair. In particular, the presence of components of the homologous recombination machinery suggests that APBs may promote telomere elongation by facilitating the homologous recombination of telomeric templates (Nabetani et al., 2004; Stavropoulos et al., 2002). This is also supported by the requirement of the homologous recombination-associated MRN complex for APB formation (Wu et al., 2000). Furthermore, APBs are suggested to be active sites of ATM and ATR dependent DNA repair (Nabetani et al., 2004). Finally, the number of APBs increases in G2 phase of the cell cycle when recombination is mainly active (Grobelny et al., 2000). We have shown that infection of normal and malignant B lymphocytes with the human oncogenic herpesvirus Epstein-Barr virus (EBV) is associated with the induction of APBs and with numerous signs of chromosomal and genomic instability (Kamranvar et al., 2007; Kamranvar and Masucci, 2011; Kamranvar et al., 2013).
Here we describe a method for detection of APBs in human B-lymphocytes. The method can be applied with minor modifications to different cell types including adherent, suspension and primary cells.
Materials and Reagents
- Cells (suspension cells)
- Formaldehyde (Merck KGaA, catalog number: K43634203 228 )
- Triton X-100 (Sigma-Aldrich, catalog number: T9284 )
- BSA (Sigma-Aldrich, catalog number: A7906 )
- Blocking reagent (Roche Diagnostics, catalog number: 11096176001 )
- Maleic acid (Sigma-Aldrich, catalog number: M0375 )
- Deionized formamide (Merck KGaA, catalog number: K25761484 902 )
- Tris-HCl
- Green-fluorescent Alexa Fluor® 488 (Life Technologies, InvitrogenTM, catalog number: A-11034 ) or Red-fluorescent Alexa Fluor® 594 (Life Technologies, InvitrogenTM, catalog number: A-11005 )
- Ethanol (Kemetyl, catalog number: 200-578-6 )
- Cy3-TelG (PANAGENE, catalog number: F1006 )
- FITC-TelC (PANAGENE, catalog number: F1009 )
- DAPI (Vector Laboratories)
- Fixation buffer (see Recipes)
- Permeabilization buffer (see Recipes)
- IF blocking buffer (see Recipes)
- FISH blocking solution (see Recipes)
- Maleic acid buffer (see Recipes)
- PNA probes (see Recipes)
- Hybridizing solution (see Recipes)
- Washing solution (see Recipes)
- PML antibody (see Recipes)
- Secondary antibody (see Recipes)
- Dehydration solution (see Recipes)
- Mounting medium (see Recipes)
Equipment
- Microscope glass slide (76 x 26 mm)
- Coverslip (preferably circular 19 mm diameter)
- Cytospin or slide centrifuge (Cytospin3 SHANDON)
- Cytospin funnel with white filter card
- Metal holder
- Hydrophobic barrier pen (ImmEdge Pen, model: H-4000 )
- Hot plate preheated to a temperature 80 °C
- Coplin jar
- Moist chamber
Procedure
- Wash the suspension cells once with PBS and prepare a dilution of 0.5 x 106 cells per ml of PBS at RT.
- Pre-label the slides and make a small hydrophobic barrier circle (~1.3 cm diameter) with ImmEdge pen around the area where the cells will be placed and let it dry before cytospin. (Hydrophobic circle will be used to fill with the buffers for fixation, permeabilization and blocking buffer.)
- Prepare the slides mounted with the filter card and cytospin funnel in the metal holder as shown in the Figure 1.
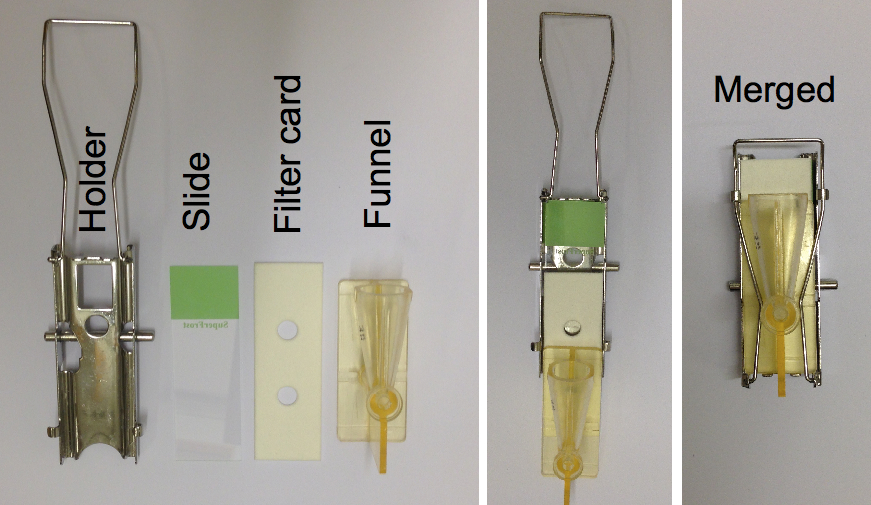
Figure 1. The figure illustrates the preparation of a slide for cytospin - Load up to 100 µl of the prepared cells in each funnel and spin for 1 min at 600 rpm.
Note: If the method will be applied to growing adherent cells on coverslip, the steps 1-4 can be skipped. - Remove the slides from the holders and let the moisture of the cells dry out for a few sec before fixation with the fixation buffer for 10 min at RT.
- Wash the cells 2 x 5 min each with PBS.
- Permeabilize the cells with permeabilization buffer for 5 min at RT.
- Remove the permeabilization buffer and incubate the cells for one hour with the IF blocking buffer in moist chamber.
- Remove the IF blocking buffer and incubate the cells in moist chamber with the fresh IF blocking buffer containing PML antibody for 2 h at RT or overnight at 4 °C.
- Wash the cells gently 3 x 10 min each with PBS on a shaker. (Washing steps can be done in any glass or plastic microscope slide staining Jars.)
Note: The slides should be protected from light in the next following steps. - Incubate the cells in moist chamber for 30 min with the secondary antibody.
- Wash the cells gently 3 x 10 min each with PBS on a shaker.
- Re-fix the cells for 5 min with fixation buffer at RT.
- Wash the cells 3 x 10 min each in PBS.
- Dehydrate the cells in coplinjar containing ethanol series, consecutively 70%, 95%, 100% EtOH, for 5 min each.
- Aspirate the ethanol completely and let the slides dry for a couple of minutes in the dark.
- Place a drop of hybridizing solution containing the PNA probe on each slide and cover it with a coverslip.
- Denature the probes in hybridizing solution for 10 min at 80 °C by placing the slides on a pre-warmed hot plate.
- Incubate the slides in moist chamber for 2 h at RT.
- Wash the slides 2 x 15 min in washing solution.
- Wash the slides 3 x 5 min in PBS.
- Air-dry the slides at RT for 10 min.
- Mount the slides with one drop of Vectashield and cover with a coverslip.
- Seal the coverslip edges with clear nail polish and let it dry.
- Keep the slides at 4 °C in dark until microscopy.
- Visualize the APBs using a confocal microscope as fluorescence signals from PML bodies co-localized with telomere signals.
- A Cell containing two or more APBs can be scored as a ALT positive cell (Figure 2).
Representative data
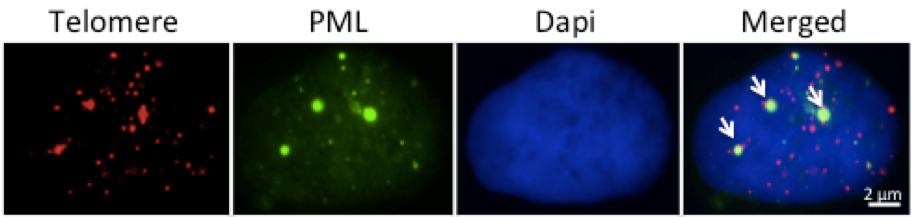
Figure 2. Representative micrograph illustrating APBs positive U2OS cell in interphase. The arrows show three APB foci in which the green PML signal co-localizes with the red telomere signal. In some cell lines the PML antibody gives a weak diffuse fluorescent background that is easily distinguished from the bright fluorescence of the PML bodies.
Notes
- Rapidly proliferating cancer cells often have shorter telomeres and higher protein expression compared to slow growing primary cells. This may cause high background in immunofluorescence and weak telomere signals in FISH. To improve the method, the incubation time with the primary PML antibody should be reduced and less photobleaching telomeric probes should be used. Cy3-TelG usually last longer than FITC-TelC and Alexa Fluor® 488 produces less background than Alexa Fluor® 594. In cells with short telomeres and high background signals, Cy3-TelG and Alexa Fluor® 488 are recommended.
Recipes
- Fixation buffer
3.7% formaldehyde in PBS (10-fold dilution of 37% commercial stock solution)
Adjust the PH to 7.5
The fixative should be freshly prepared for each experiment - Permeabilization buffer
0.5% Triton X-100 in PBS - IF blocking buffer
1% BSA and 0.1% Triton X-100 in PBS - FISH blocking solution
10% blocking reagent in maleic acid buffer: 10 g of blocking reagent should be dissolved in 100 ml of maleic acid buffer
Adjust the pH to 7.5 with NaOH and store at 4 °C - Maleic acid buffer
100 mM maleic acid and 150 mM NaCl in dH2O - PNA probes
Cy3-TelG: Cy3-OO-TTAGGGTTAGGGTTAGGG 3’
Stock concentration: 53 µM in dH2O
FITC-TelC: FITC-OO-CCCTAACCCTAACCCTAA 3’
Stock concentration: 111 µM in dH2O
Store the stocks at 4 °C in the dark - Hybridizing solution
70% deionized formamide
0.5% FISH blocking reagent (from 10% stock)
10 mM Tris-HCl (pH 7.2)
PNA telomeric probe: FITC-TelC (1:500-1,000 dilution from stock) or Cy3-TelG (1:3,000-5,000 dilution from stock)
Prepare it freshly and protect it from light - Washing solution
70% formamide
10 mM Tris-HCl (pH 7.2) in dH2O - PML antibody
Use 1:100-200 dilution in the IF blocking buffer
Monoclonal or polyclonal PML Antibodies can be used - Secondary antibody
Green-fluorescent Alexa Fluor® 488 or red-fluorescent Alexa Fluor® 594 diluted 1:1,000 in IF blocking buffer - Dehydration solution
Ethanol series 70%, 95% and 100% - Mounting medium
Vectashield embedding medium containing DAPI
Acknowledgments
Some principles of the described protocol have been extracted from the protocols of Dr Titia de Lange`s Lab. This work was supported by grants awarded by the Swedish Cancer Society and the Karolinska Institute, Stockholm, Sweden.
References
- Collado, M., Blasco, M. A. and Serrano, M. (2007). Cellular senescence in cancer and aging. Cell 130(2): 223-233.
- Grobelny, J. V., Godwin, A. K. and Broccoli, D. (2000). ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J Cell Sci 113 Pt 24: 4577-4585.
- Heaphy, C. M., Subhawong, A. P., Hong, S. M., Goggins, M. G., Montgomery, E. A., Gabrielson, E., Netto, G. J., Epstein, J. I., Lotan, T. L., Westra, W. H., Shih Ie, M., Iacobuzio-Donahue, C. A., Maitra, A., Li, Q. K., Eberhart, C. G., Taube, J. M., Rakheja, D., Kurman, R. J., Wu, T. C., Roden, R. B., Argani, P., De Marzo, A. M., Terracciano, L., Torbenson, M. and Meeker, A. K. (2011). Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 179(4): 1608-1615.
- Johnson, J. E. and Broccoli, D. (2007). Telomere maintenance in sarcomas. Curr Opin Oncol 19(4): 377-382.
- Kamranvar, S. A. and Masucci, M. G. (2011). The Epstein-Barr virus nuclear antigen-1 promotes telomere dysfunction via induction of oxidative stress. Leukemia 25(6): 1017-1025.
- Kamranvar, S. A., Chen, X. and Masucci, M. G. (2013). Telomere dysfunction and activation of alternative lengthening of telomeres in B-lymphocytes infected by Epstein-Barr virus. Oncogene 32(49): 5522-5530.
- Kamranvar, S. A., Gruhne, B., Szeles, A. and Masucci, M. G. (2007). Epstein-Barr virus promotes genomic instability in Burkitt's lymphoma. Oncogene 26(35): 5115-5123.
- Lang, M., Jegou, T., Chung, I., Richter, K., Munch, S., Udvarhelyi, A., Cremer, C., Hemmerich, P., Engelhardt, J., Hell, S. W. and Rippe, K. (2010). Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci 123(Pt 3): 392-400.
- Nabetani, A., Yokoyama, O. and Ishikawa, F. (2004). Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J Biol Chem 279(24): 25849-25857.
- Stavropoulos, D. J., Bradshaw, P. S., Li, X., Pasic, I., Truong, K., Ikura, M., Ungrin, M. and Meyn, M. S. (2002). The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet 11(25): 3135-3144.
- Wu, G., Lee, W.-H. and Chen, P.-L. (2000). NBS1 and TRF1 Colocalize at Promyelocytic Leukemia Bodies during Late S/G2 Phases in Immortalized Telomerase-negative Cells IMPLICATION OF NBS1 IN ALTERNATIVE LENGTHENING OF TELOMERES. J Biol Chem 275(39): 30618-30622.
- Yeager, T. R., Neumann, A. A., Englezou, A., Huschtscha, L. I., Noble, J. R. and Reddel, R. R. (1999). Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59(17): 4175-4179.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kamranvar, S. A. and Masucci, M. G. (2014). Detection of ALT Associated Promyelocytic Leukemia Nuclear Bodies (APBs) by Immunofluorescence-FISH (IF-FISH). Bio-protocol 4(23): e1303. DOI: 10.21769/BioProtoc.1303.
Category
Cancer Biology > Proliferative signaling > Biochemical assays > Protein analysis
Cancer Biology > Replicative immortality > Biochemical assays > DNA structure and alterations
Cell Biology > Cell staining > Nucleic acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











