- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assay of Ornithine Decarboxylase and Spermidine/Spermine N1-acetyltransferase Activities
Published: Vol 4, Iss 22, Nov 20, 2014 DOI: 10.21769/BioProtoc.1301 Views: 10285
Reviewed by: Manuel D. GaheteRenate WeizbauerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2282 Views

Reliable and Sensitive Detection of Carbonylated Proteins by Oxime Blot
Filip Luka Mikulić [...] Mladen Merćep
Aug 5, 2025 1242 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
The polyamines, spermidine (Spd) and spermine, and their diamine precursor putrescine, are important regulators of various cellular functions, such as proliferation and differentiation. Polyamine homeostasis is tightly regulated on the level of uptake, excretion, biosynthesis, interconversion and terminal catabolism. The rate-controlling enzymes of polyamine biosynthesis and interconversion are ornithine decarboxylase (ODC) and spermidine/spermine N1-acetyltransferase (SSAT), respectively. Here, we describe a protocol to assay ODC (Jänne and Williams-Ashman, 1971) and SSAT (Libby, 1978) activities from cell or tissue samples.
Keywords: 14C-L-ornithineMaterials and Reagents
- EDTA (Sigma-Aldrich, catalog number: E9884 )
- Tris-HCl (Sigma-Aldrich, catalog number: T5941 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- DL-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815 )
- Protease inhibitor cocktail, such as Complete EDTA-free protease inhibitor tablets (Roche Diagnostics, catalog number: 11873580001 )
- L-ornithine monohydrochloride (Sigma-Aldrich, catalog number: O2375 )
- Acid-treated [14C]-L-ornithine (100 µCi/ml, 40-60 mCi/mmol) (PerkinElmer, catalog number: NEC710050UC )
- Pyridoxal 5’phosphate monohydrate (PLP) (Sigma-Aldrich, catalog number: 82870 )
- Citric acid (Sigma-Aldrich, catalog number: C0759 )
- SOLVABLE (PerkinElmer, catalog number: 6NE9100 )
- [Acetyl-1-14C]-Acetyl Coenzyme A (AcCoA) (20 µCi/ml, 40-60 mCi/mmol) (PerkinElmer, catalog number: NEC313050UC ) (Note 1)
- Spermidine trihydrochloride (Sigma-Aldrich, catalog number: S2501 )
- Hydroxylamine hydrochloride (Sigma-Aldrich, catalog number: 159417 )
- ddH2O
- 96% ethanol such as ETAX A (Altia Oyj, catalog number: 12210143 )
- Liquid scintillation cocktail, such as OptiPhase HiSafe 2 (PerkinElmer, catalog number: 1200-436 )
- Buffer A (see Recipes)
- 1 M Tris-HCl (see Recipes)
- 200 mM EDTA (pH 8.0) (see Recipes)
- 100 mM DTT (see Recipes)
- Acid-treated [14C]-L-ornithine (see Recipes)
- 10 mM or 25 mM L-ornithine (see Recipes)
- 20 mM PLP (see Recipes)
- 2 M citric acid (see Recipes)
- 100 mM Spd (see Recipes)
- 1 M hydroxylamine (see Recipes)
Equipment
- 10-ml glass tubes with rubber caps (Note 2)
- Tube rack for glass tubes
- WhatmanTM 3MM Chr chromatography paper (Thermo Fisher Scientific, catalog number: 05-713-336 )
Note: Cut into 3 cm x 2 cm size and folded 4 times (Figure 1A).
- Long 18-21G needle
- 10-ml syringe
- Forceps
- Tissue homogenizer, such as 3-ml Potter-Elvehjem glass tube and pestle (Sigma-Aldrich, catalog number: P7734 ) and a drill to operate the pestle
- 0.5-ml microcentrifuge tubes
- Microcentrifuge
- Thermal cycler with block fitted for 0.5 ml tubes
- WhatmanTM Grade P81 Ion Exchange Cellulose Chromatography Paper (Thermo Fisher Scientific, catalog number: 05-171-2A ) (cut into 1.5 x 1.5 cm size)
- ParafilmTM (Sigma-Aldrich, catalog number: P7793 )
- Horizontal shaker
- A sheet of filter paper
- Wallac 4-ml plastic scintillation vials and caps (PerkinElmer, catalog number: 1200-421 )
- Liquid scintillation counter and plates fitted for 4-ml vials
Procedure
- Sample preparation
- Cell or tissue samples
- Cell samples
Resuspend cell pellets to ice-cold buffer A by pipetting up-and-down until cell suspension is homogenous; keep the samples on ice (Note 3, Note 4).
- Tissue samples
Weigh tissue and homogenize it on ice to 2-3x vol of buffer A by using Potter-Elvehjem homogenizer or other suitable homogenizer (Notes 3-4). For example, use 100 mg tissue and 200-300 µl of buffer A. Homogenize the sample until no tissue clumps remain. Transfer the homogenate to clean microcentrifuge tube with pipette.
- Cell samples
- Incubate tubes for 20 min on ice.
- Centrifuge at 15,000 x g for 30 min at +4 °C.
- Transfer the supernatant to a clean microcentrifuge tube and discard the pellet. Keep the samples on ice.
- Cell or tissue samples
- ODC activity assay
- Calculate the amount of tubes and reaction mixture needed. Each sample should be assayed in duplicate. Include also two blank reactions and two “total counts” (for determing the total radioactivity in the reaction mixture).
- Label the needed amount of glass tubes and assemble them into the tube rack. Place the rack on ice-waterbath.
- Prepare reaction mixture (Table 1) on ice.
Table 1. Reaction mixture (one reaction) for ODC activity assayComponent
Amount
1 M Tris-HCl (pH 7.4)
25 µl
200 mM EDTA
5 µl
100 mM DTT
10 µl
10 mM or 25 mM L-ornithine (Note 5)
10 µl
100 µCi/ml [14C]-L-ornithine
2 µl
20 mM PLP
5 µl
ddH2O
143 µl
Total
200 µl
- Pipet 200 µl of reaction mixture carefully to the bottom of the test tube, taking care not to touch the walls of the tubes with pipet tip (Note 6).
- Add 50 µl of sample or blank (buffer A) to the bottom of the tube.
- Pipet 25 µl of SOLVABLE onto 3 MM folded paper (Note 7), put it to the top of a glass tube and close with rubber cap (Figure 1B).
- Incubate tubes (upright) at +37 °C water bath with gentle rocking for 30 min (tissue samples) or 60 min (cell samples).
- Take the tube rack back on ice-waterbath.
- In the fume hood, add 1 ml of 2 M citric acid to the wall of the tube by using long 18-21G needle attached to a 10-ml syringe (Note 2, Note 8) (Figure 1C, D).
- Continue incubation at +37 °C water bath with gentle rocking for 15 min.
- Put the folded papers into 4-ml scintillation vials using forceps (see also Figure 2C).
- Include also two “total count” vials-pipet 100 µl of reaction mixture to a folded paper and put it into a scintillation vial.
- Add 3 ml of scintillation cocktail to each vial and close with caps (see also Figure 2D-E).
- Measure [14C]-radioactivity with liquid scintillation counter (Note 9).
- Calculating results (see also Table 3):
- Check the label of [14C]-L-ornithine for specific activity and calculate the total molar amount of L-ornithine (labelled + unlabelled) in the reaction mixture (200 µl):
Labelled L-ornithine:
clabelled = radioactive c (mCi/ml)/specific activity (mCi/mmol)
nlabelled = clabelled*Vlabelled
ntotal = nlabelled+nunlabelled
- Next calculate how many cpm correspond to one pmol of L-ornithine:
Relative specific activity (RSA) (pmol/cpm) = ntotal/[(average “total count” cpm)*2]
- Then calculate the result:
- For cells
[(average sample cpm - average blank cpm) * RSA]/(mg protein per 50 µl sample) = pmol/h/mg protein
- For tissue
{[(average sample cpm - average blank cpm) * RSA]/(mg protein per 50 µl sample)}/0.5 h = pmol/h/mg protein
Activity can be expressed as per mg protein, mill. cells, µg DNA or mg tissue.
- For cells
- Check the label of [14C]-L-ornithine for specific activity and calculate the total molar amount of L-ornithine (labelled + unlabelled) in the reaction mixture (200 µl):
- For tissue and cell samples, dilution of the sample is generally needed when cpm-values exceed ~15,000.
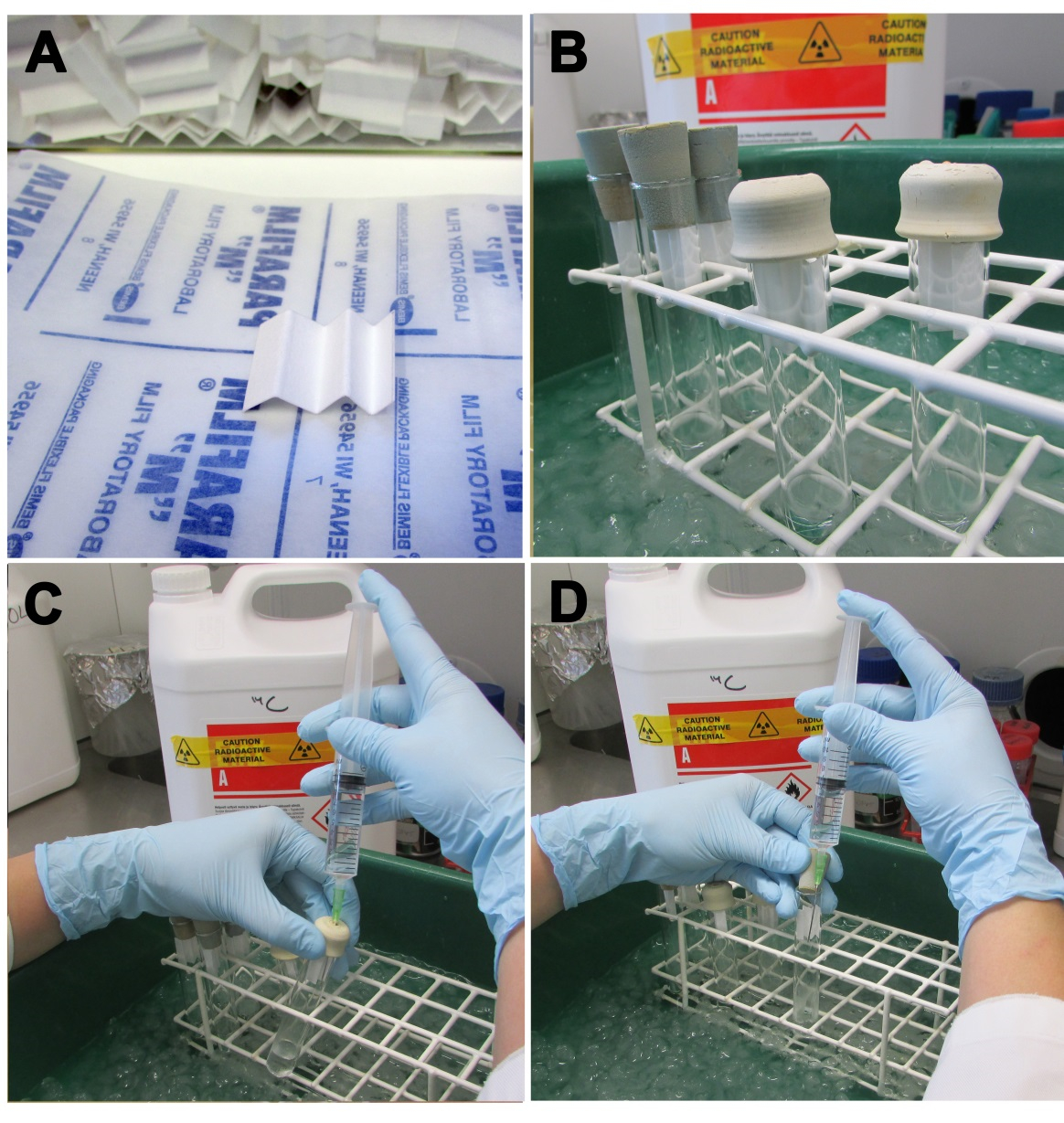
Figure 1. ODC activity assay. A. Folded Whatman 3 MM filter papers. B. ODC reaction tubes with folded papers in place. C. Stopping the reaction with the addition of 2 M citric acid, using needle-penetrable rubber caps and D normal rubber caps.
- Calculate the amount of tubes and reaction mixture needed. Each sample should be assayed in duplicate. Include also two blank reactions and two “total counts” (for determing the total radioactivity in the reaction mixture).
- SSAT activity assay
- Calculate the amount of tubes and reaction mixture needed. Each sample should be assayed in duplicate. Include also two blank reactions and two “total counts” for determing the total radioactivity in the reaction mixture.
- Label the needed amount of 0.5-ml microcentrifuge tubes and place them on ice.
- Prepare reaction mixture (Table 2) on ice.
Table 2. Reaction mixture (one reaction) for SSAT activity assay
Component
Amount
1 M Tris-HCl (pH 7.8)
10 µl
100 mM Spd
3 µl
100 mM DTT
1 µl
(20 µCi/ml) [14C]-AcCoA
2.5 µl
ddH2O
73.3 µl
Total
90 µl
- Pipet 90 µl of reaction mixture to each tube.
- Pipet 10 µl of sample or blank (buffer A) to each tube.
- Incubate in a thermal cycler for 10 min at +37 °C (Note 10).
- Cool to +4 °C and add 20 µl of 1 M hydroxylamine.
- Heat at +100 °C for 3 min.
- Centrifuge tubes at 15,000 x g for 5 min at RT.
- Place P81 paper squares onto Parafilm and number them with pencil (Figure 2A).
- Pipet 40 µl of sample supernatant to each paper square and let dry at RT. Include also two “total counts”- pipet 45 µl of reaction mixture to filter paper.
- Place the “total count” paper squares into 4-ml scintillation vials using forceps, add 3 ml of scintillation cocktail to each vial and close them with caps (Figure 2C-E).
- Put sample and blank paper squares to 1 L Erlenmeyer flask.
- Wash paper squares with dH2O for 3 x 5 min and 1 x 10 min (500 ml each wash), shaking on a horizontal shaker at ~200 rpm (Notes 11-12) (Figure 2B).
- Wash paper squares once with 400 ml of 96% ethanol for 5 min, shaking on a horizontal shaker at ~200 rpm (Note 11).
- Pour paper squares with ethanol to a large container, pick them with forceps onto a big sheet of filter paper and let dry.
- Put the paper squares into 4-ml scintillation vials (Figure 2C).
- Add 3 ml of scintillation cocktail into each vial and close them with caps (Figure 2D-E).
- Measure [14C]-radioactivity with liquid scintillation counter (Note 9).
- Calculating the results (see also Table 3):
- Check the [14C]-AcCoA batch label for specific activity and calculate its molar amount in the reaction mixture (90 µl):
c = radioactive c (mCi/ml)/specific activity (mCi/mmol)
n = c * V
- Next calculate how many cpm correspond to one pmol of AcCoA:
Relative specific activity (RSA) (pmol/cpm) = n/[(average “total count” cpm)*2]
- Then calculate the result:
{[(average sample cpm - average blank cpm) * RSA] * (120 µl/40 µl)/(mg per 10 µl sample)} * 6 = pmol/h/mg
Activity can be expressed as per mg protein, mill. cells, µg DNA or mg tissue.
- Check the [14C]-AcCoA batch label for specific activity and calculate its molar amount in the reaction mixture (90 µl):
- For tissue and cell samples, dilution of the sample is generally needed when cpm-values exceed ~6,000.
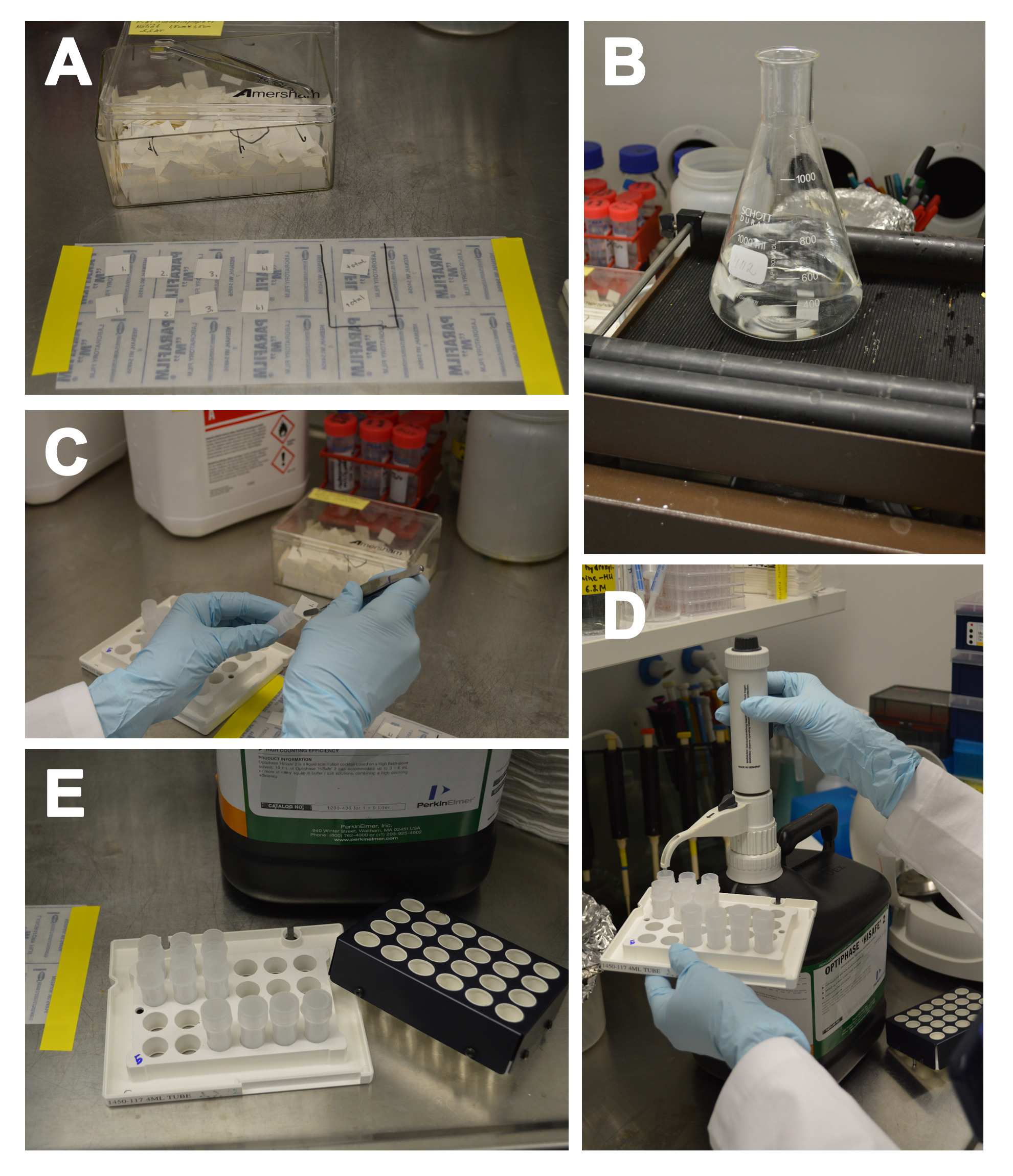
Figure 2. SSAT activity assay and preparing the samples for liquid scintillation counting. A. Numbered P81 paper squares for samples, blanks and total counts. B. Washing the paper squares. C. Placing the paper squares to liquid scintillation vials. D. Adding scintillation cocktail with dispenser. E. Capped and uncapped vials and the cassette cover.
- Calculate the amount of tubes and reaction mixture needed. Each sample should be assayed in duplicate. Include also two blank reactions and two “total counts” for determing the total radioactivity in the reaction mixture.
Representative data
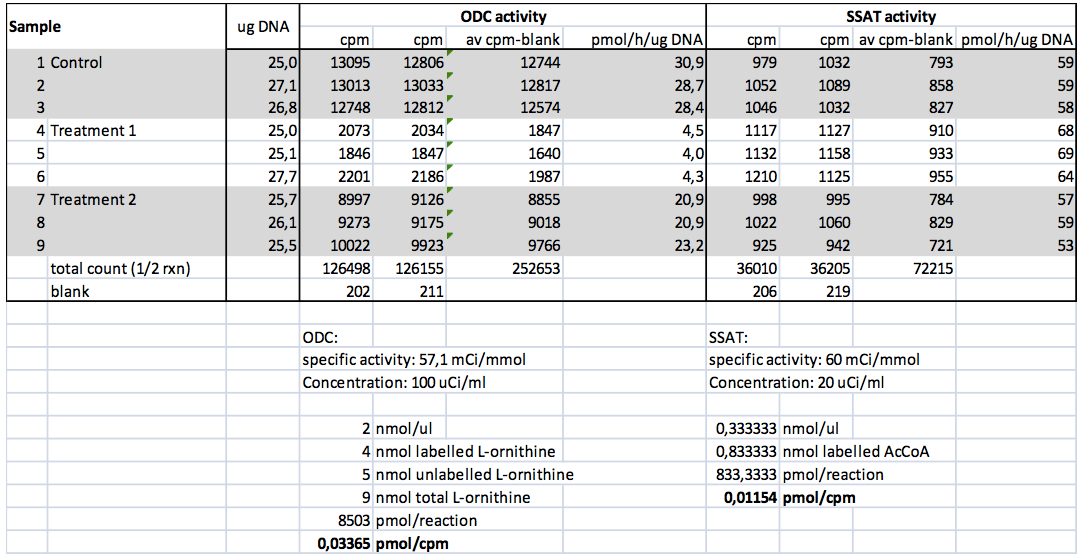
Table 3. Representative data from an experiment with DU145 cells
Notes
- AcCoA is very labile. After thawing the package, aliquot and store at -20 °C.
- Using needle-penetrable rubber caps is recommended to avoid any possible release of radioactive CO2 from the tube.
- The cells/tissue can be either fresh or frozen at -70 °C.
- The amount of buffer used depends on the ODC/SSAT activity in the particular cell/tissue type. The needed amount of cleared lysate is 20 µl for SSAT assay and 100 µl for ODC assay. We recommend using at least 40 mg of tissue and at least 0.5 mill. cells/200 µl buffer.
- Use 10 mM L-ornithine for cell samples and 25 mM L-ornithine for tissue samples.
- Extra-long pipette tips are convenient when pipetting to the bottom of the tubes. Unexpectedly high cpm-values are usually caused by contaminating reaction mixture on the wall of the glass tube.
- SOLVABLE absorbs released [14C]-CO2.
- Do not touch the paper with the needle, otherwise bound [14C]-CO2 will be released. If using normal rubber caps, open it just slightly in order to fit the needle into the tube and close the cap immediately after the addition of citric acid.
- For optimal results, leave vials o/n at RT before measurement.
- Reactions are conveniently done in a thermal cycler, but can also be done on water baths or heat blocks.
- Shaking speed depends on the size of the shaker. Use speed that enables the paper squares to gently circle around the flask. Too high speed will lead to loss of paper from the square corners.
- High blank values (>500 cpm) indicate insufficient washing or too high amount of samples in the same flask. Either use two flasks to wash the paper squares, or add a couple of additional 5 min washes with H2O.
- Triton X-100 is not absolutely necessary for tissue samples, but it enhances cell breakage.
- Commercial preparations of [14C]-L-ornithine contain variable amounts of radioactive CO2, which will result in high blank values. Acid treatment removes any residual radioactive CO2 (Jänne and Williams-Ahsman, 1971).
Recipes
- Buffer A (Note 13)
Mix 1.25 ml of Tris-HCl pH 7.4, 250 µl of 200 mM EDTA pH 8.0, 50 µl of Triton X-100 and 500 µl of 100 mM DTT
Add ddH2O to final volume of 50 ml
Dissolve one Complete EDTA-free tablet to the solution
Aliquot and store at -20 °C
- 1 M Tris-HCl (pH 7.4 or pH 7.8)
Dissolve 157.6 g of Tris-HCl to ~800 ml of ddH2O
Adjust pH to 7.4 or 7.8 with NaOH
Add ddH2O to final volume of 1,000 ml
Filter-sterilize (0.2 µm)
Stored at RT
- 200 mM EDTA (pH 8.0)
Weigh 58.448 g of EDTA to ~800 ml of ddH2O
Adjust pH slowly to 8.0 with NaOH - EDTA dissolves when pH approaches 8
Add ddH2O to final volume of 1,000 ml
Filter-sterilize (0.2 µm)
Stored at RT
- 100 mM DTT
Dissolve 1.5425 g of DTT to ddH2O in a final volume of 100 ml in the fume hood
Filter-sterilize (0.2 µm)
Aliquot and stored at -20 °C
- Acid-treated [14C]-L-ornithine (Note 14)
Mix 0.5 ml of [14C]-L-ornithine with 1 ml of 0.1 M HCl
Evaporate to dryness in a rotary evaporator in a fume hood
Dissolve residue into 0.5 ml of 0.01 M HCl
Aliquot and stored at -20 °C
- 10 mM or 25 mM L-ornithine
Dissolve 168.62 mg or 421.55 mg of L-ornithine monohydrochloride to ddH2O in a final volume of 100 ml to obtain 10 mM or 25 mM concentration, respectively
Filter-sterilize (0.2 µm)
Aliquot and stored at -20 °C
- 20 mM PLP
Dissolve 530.32 mg of pyridoxal 5’phosphate monohydrate to ddH2O to a final volume of 100 ml
Filter-sterilize (0.2 µm)
Aliquot and stored at -20 °C
- 2 M citric acid
Dissolve 384.24 g of citric acid to ddH2O to a final volume of 1,000 ml
Filter-sterilize (0.2 µm)
Stored at +4 °C
- 100 mM Spd
Dissolve 2.546 g of Spd to ddH2O to a final volume of 100 ml
Filter-sterilize (0.2 µm)
Aliquot and stored at -20 °C
- 1 M hydroxylamine
Dissolve 6.949 g of hydroxylamine hydrochloride to ddH2O to a final volume of 100 ml
Filter-sterilize (0.2 µm)
Stored at RT
Acknowledgments
This work was supported by the strategic funding of the University of Eastern Finland and the Academy of Finland. The protocols were modified from the original versions developed by Jänne and Williams-Ashman (1971) and Libby (1978).
References
- Jänne, J. and Williams-Ashman, H. G. (1971). On the purification of L-ornithine decarboxylase from rat prostate and effects of thiol compounds on the enzyme. J Biol Chem 246(6): 1725-1732.
- Libby, P. R. (1978). Calf liver nuclear N-acetyltransferases. Purification and properties of two enzymes with both spermidine acetyltransferase and histone acetyltransferase activities. J Biol Chem 253(1): 233-237.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hyvönen, M. T., Keinänen, T. and Alhonen, L. (2014). Assay of Ornithine Decarboxylase and Spermidine/Spermine N1-acetyltransferase Activities. Bio-protocol 4(22): e1301. DOI: 10.21769/BioProtoc.1301.
Category
Cell Biology > Cell metabolism > Other compound
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









