- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Severe Fever with Thrombocytopenia Syndrome Virus Infection of Cell Cultures
Published: Vol 4, Iss 20, Oct 20, 2014 DOI: 10.21769/BioProtoc.1274 Views: 8869
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Drug Susceptibility Assay for HBV Using Southern Blotting
Sung Hyun Ahn [...] Kyun-Hwan Kim
Apr 20, 2015 11334 Views

Isolation of Exosomes from Semen for in vitro Uptake and HIV-1 Infection Assays
Marisa N. Madison [...] Chioma M. Okeoma
Apr 5, 2017 12857 Views

Sleeping Beauty Transposon-based System for Rapid Generation of HBV-replicating Stable Cell Lines
Jin-Wei Zheng [...] Quan Yuan
Jul 5, 2018 9725 Views
Abstract
Severe fever with thrombocytopenia syndrome virus (SFTSV) can infect multiple cells, such as Vero, RD, BHK21, HUVEC and 293T. Vero cells are highly susceptible, so they were used to isolate virus from sera of patients, as well as for producing virus in the laboratory. Other cells can be used for additional functional analysis.
Materials and Reagents
- Vero cells (acquired from ATCC, CCL-81 )
- SFTSV from patients sera
- Dulbecco’s modified Eagle medium (DMEM) + 4.5 g/L D-glucose with L-Glutamine (Life Technology, catalog number: C11965500BT )
- Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10091148 )
- Penicillin streptomycin (PS) (Life Technologies, Gibco®, catalog number: 15140-122 )
- Antibody against nuclear protein (NP) of SFTSV
Note: Produced in rabbit by the Antibody Facility of our institute (Sun et al., 2014), the antibody was affinity purified, 1 mg/ml, the working concentration is 5 μg/ml.
- TRIzol reagent (Life Technologies, InvitrogenTM, catalog number: 15596-026 )
- Reverse Transcriptase M-MLV (RNase H) [Takara Biotechnology (Dalian), catalog number: D2639A ]
- SYBR Premix Ex TaqTM Perfect Real Time [Takara Biotechnology (Dalian), catalog number: DRR039A ]
- Methanol
- 4',6-diamidino-2-phenylindole (DAPI)
- Alexa Fluor 488 goat anti-rabbit (Life Technologies, InvitrogenTM, catalog number: A11008 )
- Hipure Plasmid Filter Midiprep Kit (Life Technologies, catalog number: K210005 )
- Maintaining cell culture (see Recipes)
- Infection cell culture (see Recipes)
- Mounting media (see Recipes)
Equipment
- T25 flask (25 cm2) (BD, FalconTM, catalog number: 353108 )
- Tissue culture plate, 24-well flat bottom (BD, FalconTM, catalog number: 353047 ) or 48-well plate (BD, FalconTM, catalog number: 353078 )
- 25-cm bottles
- 37 °C, 5% CO2 incubator
- 7500 fast Real Time PCR System (Life Technologies, Applied Biosystems®)
- Zeiss LSM510 Meta laser scanning confocal microscope
Procedure
- Virus isolation
- Vero cells were seeded in T25 flasks with 80% confluence, cultured in 5 ml DMEM/10% FBS/PS one day earlier before being inoculated with SFTSV patient sera.
- The sera were acquired from the hospital. The vein blood was collected from patients through the clinical laboratory of the hospital and sera were acquired by centrifugation at 2,500 rpm.
- The virus in the sera of patients was first quantified by quantitive real time PCR with specific primers designed for amplifying NP of SFTSV (see below). Serum with high SFTSV concentration (about 108/ml) was used to isolate SFTSV.
- A total of 500 μl of the patient’s sera was inoculated in 4.5 ml DMEM/2% FBS in 25-cm bottles, incubated at 37 °C, 5% CO2 incubator, and changed with fresh DMEM/2% FBS in 24 h post infection.
- The sera were acquired from the hospital. The vein blood was collected from patients through the clinical laboratory of the hospital and sera were acquired by centrifugation at 2,500 rpm.
- Cultured supernatant was collected 72 h after virus inoculation of Vero cells, and the supernatant was added to newly seeded Vero cells in a T25 flask Virus.
- All collected cultured supernatant was used for virus amplification for the first two to three passages, and then 2.5 ml supernatant was used to amplify the virus.
- The virus was amplified for 3 to 5 generations with cultured supernatant till the copies of NP reached 108 to 109/μl.
- SFTSV virus in DMEM/2% FBS/PS was distributed to 1.5 ml micro-centrifuge tubes and stored at -80 °C, avoiding freeze thaw cycles, even though the virus is not that sensitive to the free-thaw process.
- All collected cultured supernatant was used for virus amplification for the first two to three passages, and then 2.5 ml supernatant was used to amplify the virus.
- Titration of virus measured by quantitative Real Time-PCR.
The supernatant was spun at 3,000 rpm for 5 min. 200 μl cultured supernatant was added to 1 ml TRIzol reagent.- Total RNA was isolated from supernatant using TRIzol reagent according to the manufacturer’ instructions. 2 μg of total RNA was used to synthesize cDNA using Reverse Transcriptase M-MLV (RNase H) in 20 μl reaction following the user manual.
- Next, 2 μl cDNA from each sample was used for quantitive real-time PCR by using SYBR Premix Ex Taq TM Perfect Real Time according to the manufacturer’s instructions. Primers (FW: AGCAGCTCAATTTGACTGAG, RW: TCATCTCCACCTGTCTCCTT) were designed to amplify the 87 bp fragment of NP.
- The full length NP DNA was amplified from virus cDNA, and was cloned to pCI-neo Vector; the plasmid was prepared using Hipure Plasmid Filter Midiprep Kit according to the user manual.
- The concentration of the plasmid was quantified, and DNA copies were calculated according to their molecular mass. These were diluted to serial copies (101 to 107) for making the standard curve, after which the quantitative real time-PCR was performed.
20 μl of PCR reaction include:
10 μl 2x SYBR Premix ExTaq
0.4 μl each of 10 μM Forward primer and Reverse primer
0.4 μl ROX Reference Dye II
6.8 μl H2O
2 μl DNA sample
Following PCR were performed at 95 °C 10 sec, 40 repeat cycles of 95 °C 5 sec and 60 °C 30 sec, and dissociation at last.
- Total RNA was isolated from supernatant using TRIzol reagent according to the manufacturer’ instructions. 2 μg of total RNA was used to synthesize cDNA using Reverse Transcriptase M-MLV (RNase H) in 20 μl reaction following the user manual.
- The titration of the virus can also be determined by immunostaining with antibody against nuclear protein (NP) of SFTSV.
Vero or HUVEC cells were seeded in 24 well plates with 80% confluence one day earlier and infected with different amounts of SFTSV collected after 5 generations (such as 2 μl, 5 μl, 10 μl, 20 μl to 40 μl in DMEM/2% FBS) for two hours in 37 °C, 5% CO2 incubator, washed with PBS for twice in 2 h post infection and change with DMEM/2% FBS.
After one day (24 h) infection, the cells were washed with 500 μl PBS once, incubated for 2 to 5 min for every washing step with PBS, and then the cells were fixed with 500 μl 100% methanol for 10 min at RT, and then washed with 500 μl PBS once.
Next, they were incubated with 200 μl antibodies against NP (produced in rabbit) at 5 μl/ml in PBS/2% BSA at 37 °C incubator for 1 h. Afterwards, the cells were washed with 500 μl PBS three times. The cells were incubated with 2 μg/ml of Alexa Fluor conjugated Goat anti-rabbit IgG at 37 °C incubator for 1 h and then washed with PBS three times. Following that, the cells were covered with 80 μl mounting medium. Nuclei were stained using 4',6-diamidino-2-phenylindole (DAPI) in mounting media. Images were captured with fluorescence microscope at 10 or 20x lens.
- Vero cells were seeded in T25 flasks with 80% confluence, cultured in 5 ml DMEM/10% FBS/PS one day earlier before being inoculated with SFTSV patient sera.
- Virus infection in vitro
- Vero or other cells were seeded one day earlier on 24 or 48 well plates depending on the experiment. Here, 48 well plates are used as an example.
- The 10 μl virus collected from the supernatant was diluted in 200 μl DMEM/2% FBS (MOI 1) and added to the cells.
- After the cells were incubated at the 37 °C, CO2 incubator for 2 h, the cell was washed with PBS once, and then changed with DMEM/2% FBS.
- In order to test the infection of SFTSV, after 14 to 24 h, the cell culture was collected, and total RNA was prepared using TRIzol reagent, and quantitive Real Time PCR were performed as above description. Alternatively, the cells were fixed with methanol and stained with NP antibody as above.
- Vero or other cells were seeded one day earlier on 24 or 48 well plates depending on the experiment. Here, 48 well plates are used as an example.
Representative data
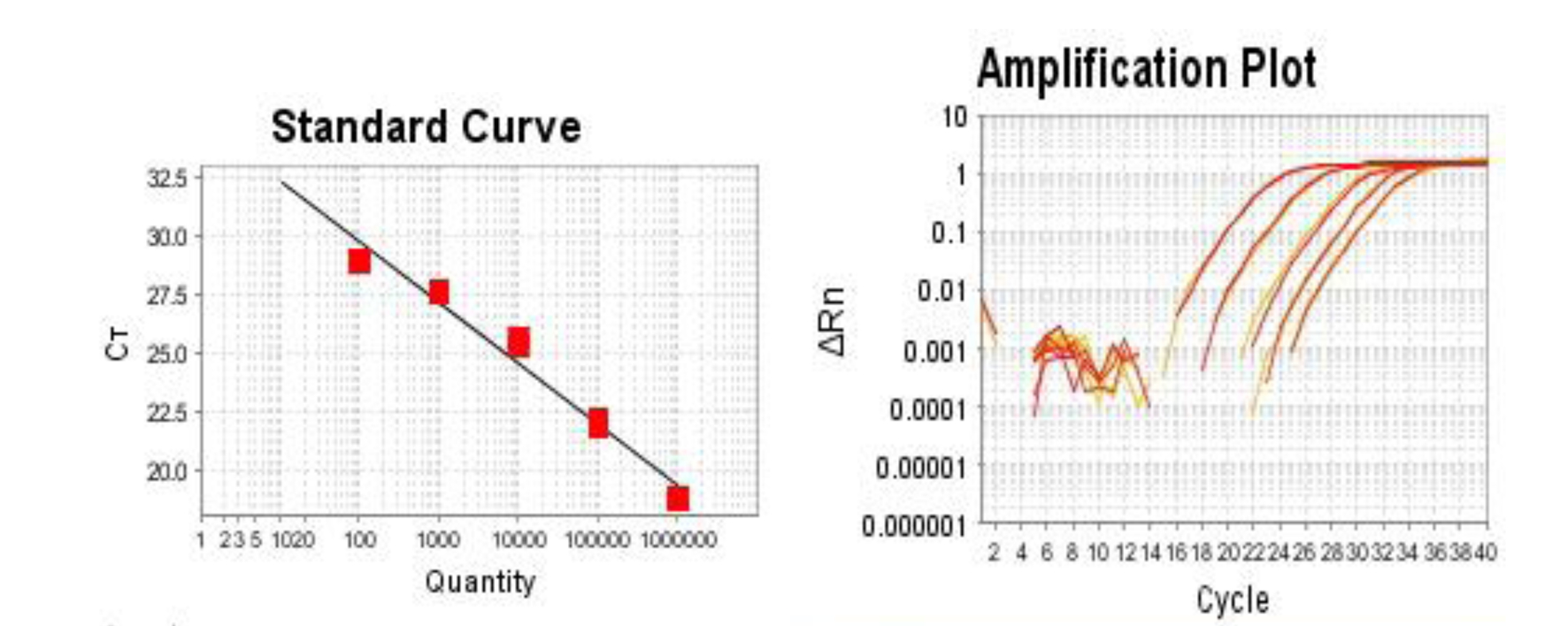
Figure 1. Standard curve of quantitative real time-PCR for SFTSV NP at 102 to 106 copies. When quantitative PCR was performed, the plasmid NP- pCI-neo was diluted from 102 to 106. Next, 2 μl was added in 20 μl reaction using SYBR Premix Ex Taq as per the user manual, the reaction was duplicated, and PCR was performed on the 7500 fast Real Time PCR System. X axis is the quantity of the NP copies. The Y axis is the value of CT of PCR at different copies. Sample copies will be automatically generated by the program depending on the standard curve. The right figure is the amplification plot.
Figure 2. HUVEC cells were infected with SFTSV virus produced in Vero cells (vero-105 batch). Cells infected with different amounts of virus (2 μl, 5 μl, 10 μl, 20 μl and 40 μl) as indicated in the figure. Cells were stained with antibodies against NP (green), nuclei were stained using 4',6-diamidino-2-phenylindole (DAPI) in mounting media (blue). Images were acquired using a Zeiss LSM510 Meta laser scanning confocal microscope at 20 times.
Notes
- When SFTSV infects different cells, the detection time for RT-PCR or immune staining will be selected according the experimental requirement when multiple cells are used. This is because the cells have different sensitivities to SFTSV infection.
Recipes
- Maintaining cell culture
DMEM/10% FBS/PS
- Infection cell culture
DMEM/2% FBS/PS: 1 ml FBS, 500 μl PS and 48.5 ml DMEM for preparing 50 ml DMEM/2% FBS/PS
- Incubate condition
37 °C 5% CO2 incubator
- Mounting media
1:1 glycerol:PBS
0.1% N-propyl gallate
Acknowledgments
We thank Zhaoling Sun from Yiyuan County Chinese Medical Hospital for providing us with sera samples from SFTSV patients. This work was supported by the Chinese Ministry of Science and Technology (2010CB530101, 2011CB812501) and the Beijing Municipal Government.
References
- Sun, Y., Qi, Y., Liu, C., Gao, W., Chen, P., Fu, L., Peng, B., Wang, H., Jing, Z., Zhong, G. and Li, W. (2014). Nonmuscle myosin heavy chain IIA is a critical factor contributing to the efficiency of early infection of severe fever with thrombocytopenia syndrome virus. J Virol 88(1): 237-248.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sun, Y. and Li, W. (2014). Severe Fever with Thrombocytopenia Syndrome Virus Infection of Cell Cultures. Bio-protocol 4(20): e1274. DOI: 10.21769/BioProtoc.1274.
Category
Microbiology > Microbe-host interactions > Virus
Microbiology > Microbe-host interactions > In vitro model > Cell line
Microbiology > Microbial cell biology > Cell isolation and culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











